Spike Tools contains tools used to process neural data (threshold detection). This repository will be mostly useful to folks with a recording rig setup similar to the one described below.
- Intan RHD Recording System: Hardware used for acquiring extracellular electrophysiology data.
- RHD Recording Controller
- SPI interface cable
- RHD 32-channel headstage
- DAQ [USB-6501, National Instruments]
- Camera: Used to monitor the animal.
- Eye-tracker [EyeLink 1000, SR Research]: Used to monitor the animal's eyes (mainly eye position).
- Photodetector [DET36A2, Thorlabs]: Used to obtain accurate timing of stimulus presentation. Each time a stimulus is presented, we simultaneously present a small white square below the photodector.
- Juice Reward System
- Juice pump (hidden from view)
- 1000 mL beaker
- Juice tube
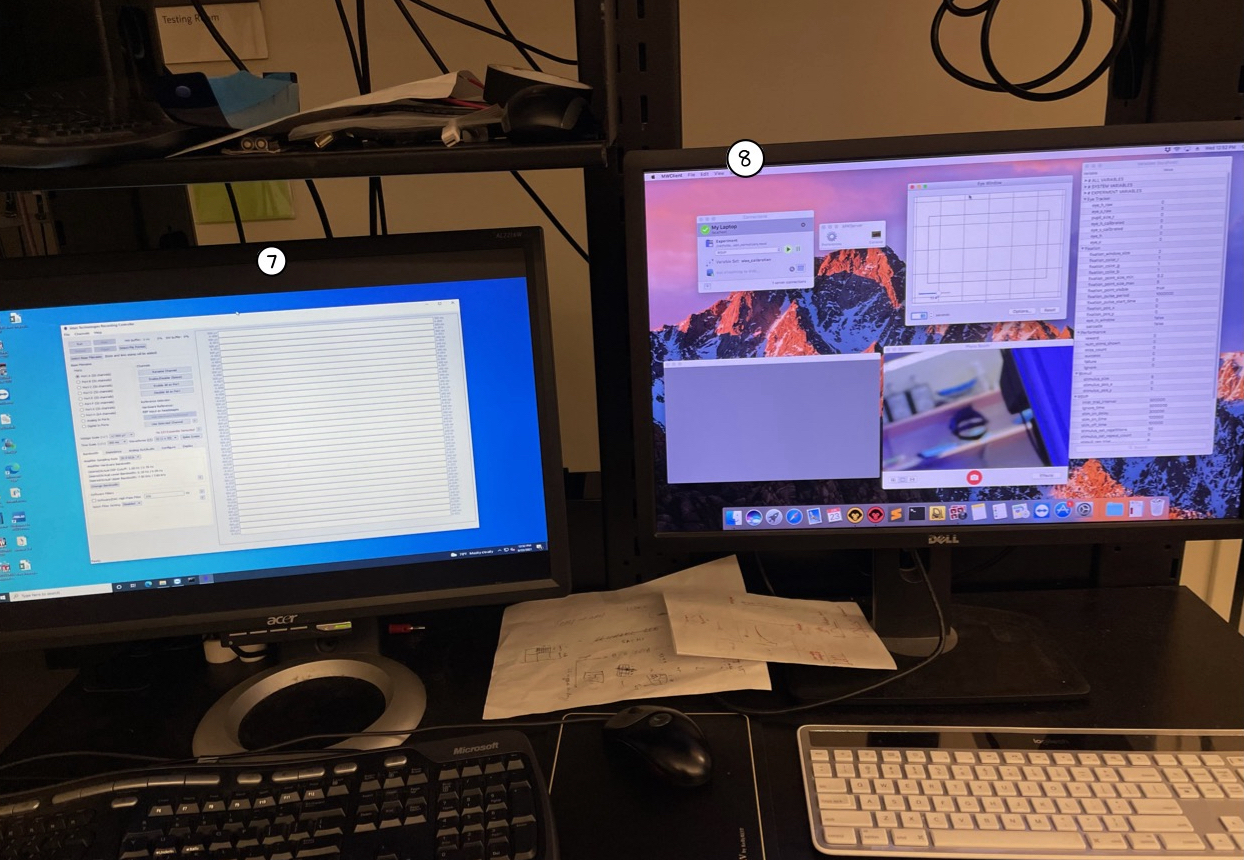
- Intan Recording Controller Software: Software used for recording extracellular electrophysiology data. This is now a legacy software (as of February 2021) -- the newer version, called RHX Acquisition, allows for both recording and stimulation. Please refer to the user manual or website for general know-hows and installation instructions.
- MWorks: Software used to design and run realtime experiments.
👉 The instructions are for the legacy software. The steps might not exactly be the same if you're using the newer versions of the software (released in 2021 or later).
-
If you don't have a pre-saved settings file, please follow the steps in the section below. Else, click on "File"->"Load Settings" to load the settings file.
-
Click "Run" to see the acquired signals in real-time. Use this time to visually inspect all channels to ensure the connections are secure.
-
Click "Select Base Filename", choose the directory where you want to save output data, and type in the base name of the output directory housing all data files. Convention has been to use
animalname_experimentname(e.g., oleo_domain-transfer). -
Click "Trigger". Double-check the trigger source channel is correct (this is the digital channel that gets a pulse when we press the green start button on MWorks). Set posttrigger buffer to 9999 seconds (max), and hit "OK". You'll see a message "Waiting for logic high trigger on digital input X..." on the left-bottom of the screen. You can now start the experiment (on MWorks). Once you start the experiment, the message on the left-bottom of the screen should switch to something-along-the-lines "Saving data at output_directory_path".
-
Click "Select File Format", and choose "One File Per Channel" format. This sets the format of the output file(s).
-
Amplifier Sampling Rate: Set it to 20.0 kS/s.
-
Software Filters: Check-mark the "Software/DAC High-Pass Filter", and set it to 400 Hz. Set the Notch Filter Setting to 60 Hz. (Note, the filtering is only for display purposes).
-
Voltage Scale: Set it to +- 100\microV.
-
Time Scale: Set it to 2000ms.
-
Waveforms: Set it to 32 (4 x 8).
-
Enable/disable all required channels by first clicking on the appropriate channel, and then the "Enable/Disable" button.
-
Click "File"->"Save Settings" to save these settings in a file which you can easily load next time.
-
Start MWorks Server and Client. If you're setting up MWorks for the first time in your current rig, read-on. Else, Skip to 3.
-
Click "Preferences" on MWorks Server. Double-check the display settings are correct. This is important for correct rendering of your stimuli.
-
Ensure your eye-tracker is on, then click the folder icon to load your experiment. (MWorks, by default, checks all I/O devices used in your experiment. You can disable this, but refer to the official MWorks documentation for this).
-
Click the icon with the three dots to load or create the eye calibration file. To load, select the file from dropdown list and hit "load". To create a new file, type in the file name and hit "create". All calibration files are saved by default to
./Document/MWorks/Experiment Storage(there's a separate folder for each experiment). -
Select "Eye Calibration" from the dropdown menu in MWorks Client, and press the green start button run the eye calibration protocol. To save the calibration, press the icon with the three dots, re-type the calibration file name, then hit "save".
-
You're now ready to start the experiment. Choose your experiment protocol from the dropdown menu in MWorks Client and press the green start button. The output data will be saved to
./Document/MWorks/Databy default.
At the end of each recording session, we transfer the data files from the local machine to a server aka braintree. This is particularly important for the neural data files, since the file sizes can get big (up to ~150 GB per day).
Intan software output goes to the appropriate intanraw sub-directory.
MWorks software output goes to the appropriate mworksraw sub-directory.
Directory structure on server aka braintree
🗂️ projects/
├─ 🗂️ domain-transfer/
├─ 🗂️ muri1320/
├─ 🗂️ normalizers/
│ ├─ 🗂️ monkeys/
│ │ ├─ 🗂️ oleo/
│ │ │ ├─ 🗂️ intanraw/
│ │ │ │ ├─ 🗂️ oleo_normalizers_210609_110927/
│ │ │ │ ├─ 🗂️ oleo_normalizers_210609_134950/
│ │ │ ├─ 🗂️ mworksraw/
│ │ │ │ ├─ 📄 oleo_normalizers_210609_110709.mwk2
│ │ │ │ ├─ 📄 oleo_normalizers_210609_134829.mwk2
│ │ │ ├─ 🗂️ intanproc/
│ │ │ ├─ 🗂️ mworksproc/
│ │ ├─ 🗂️ solo/
+--------+ +--------------------------------------------------------------------------------+
| MWorks | -- (.mwk2) --> | mwk.py <.mwk2 file> <photodector signal (analog)> <stimulus on signal (digital)> |
+--------+ +----------------------------------------+---------------------------------------+
|
|
(.csv)
|
+---------------------+ |
| Intan Technologies | |
+----------+----------+ |
| |
+-----------+-------------+ |
| | | |
(amp_0.dat) (amp_1.dat) ... (amp_n.dat) |
| | | |
v v v |
+---------------------------------------+ |
| utils.spikeutils.get_spike_times | |
| collect spike time information. | |
+-------------------+-------------------+ |
| |
+-----------+--------------+ |
| | | |
(0_spk.mat) (1_spk.mat) ... (n_spk.mat) |
| | | |
v v v |
+-------------------------------------------------------+ |
| utils.spikeutils.get_psth | |
| combine spike event times and behavioral data to | <----------+
| to make a PSTH. |
+----------------------+--------------------------------+
|
+---------------+----------------+
| | |
(0_psth.mat) (1_psth.mat) ... (n_psth.mat)
| ` | |
v v v
+-----------------------------------------------+
| utils.spikeutils.combine_channels |
| combine PSTH files for individual channels |
| into a single file. |
+----------------------+------------------------+
|
|
(session_psth.mat)
|
|
|
.
. x m sessions
.
V
+-------------------------------------------------------------+
| utils.spikeutils.combine_sessions |
| combine PSTH files for all experiment sessions by |
| concating along the repetition axis. |
+----------------------+--------------------------------------+
|
|
(experiment_psth.h5)
|
|
.
. Package data for benchmarking via brainio_collection/brainio_contrib. This involves
. writing a script that combines neural data, image metadata, and array metadata into
. a data assembly supported by Brain-Score.
.
V
(experiment_assembly.nc)