This package provides a convenient python interface to Jerome Friedman's Fortran implementation of glmnet. It provides Elastic and Logistic net models, cross validation, and plotting utilities.
It is very much a work in progress.
- David Warde-Farley (2010)
- Matthew Drury (2014)
To create an elastic net model:
from glmnet import ElasticNet
enet = ElasticNet(alpha=.1)
(Here, alpha is the relative weighting between the L1 and L2
regularization terms). The model can then be fit using a design matrix X and
a response vector y:
enet.fit(X, y)
Glmnet also accepts sparse design matricies, using the compressed sparse column format:
from scipy.sparse import csc_matrix
Xsp = csc_matrix(X)
enet.fit(Xsp, y)
After fitting, the model can be used to generate predictions on new data:
enet.predict(X')
(Note that this generates predictions for each value of lambda that was
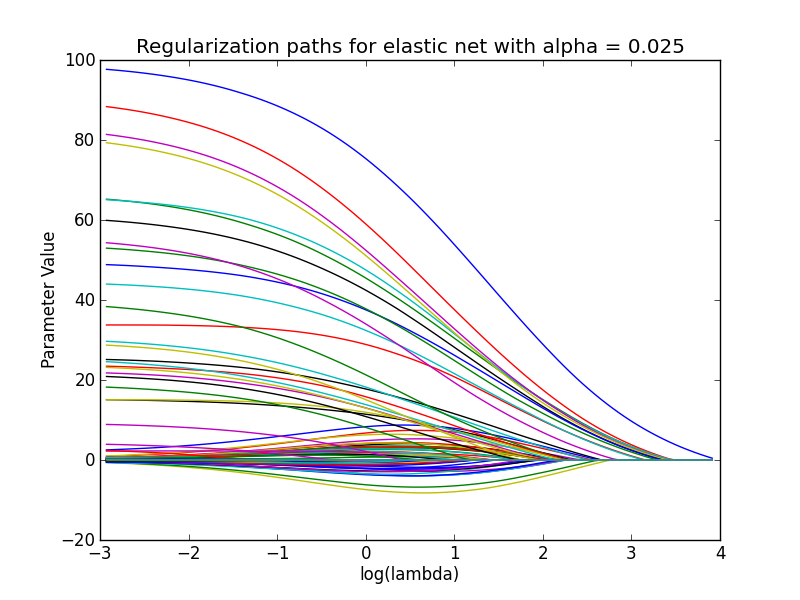
used in the coordinate descent algorithm). The parameter paths can also be
visualized, that is, the values of the model parameters for each lambda:
enet.plot_paths()
To select a value of lambda cross-validation can be used:
from glmnet import ElasticNet, CVGlmNet
enet = ElasticNet(alpha=.1)
enet_cv = CVGlmNet(enet, folds=10, n_jobs=10)
enet_cv.fit(X, y)
Glmnet then fits ten models for each value of lambda, and chooses the best
model by observing which optimizes the out of fold deviance.
Note: glmnet uses the joblib.Parallel function to parallelize its fitting
across folds, there is a known bug in some versions of OSX where using this
causes a race condition and the fitting will hang. Setting n_jobs=1 will
disable the cross validation, at the expense of fitting the models in series.
The parallelization has been tested on various linux boxes with no issues. See
this sklearn issue for more
information.
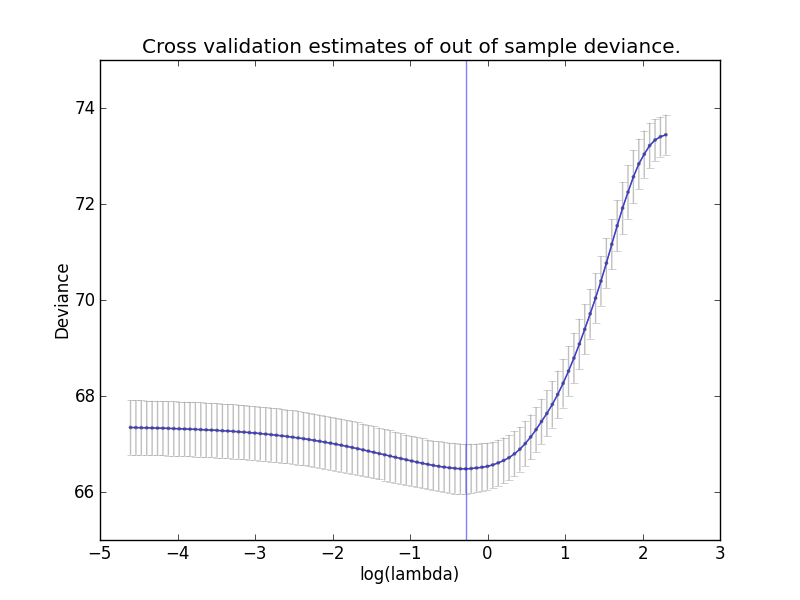
Once the cross validation is fit, the mean out of fold deviances for each value
of lambda can be viewed, along with their standard deviations:
enet_cv.plot_oof_devs()
 The cross validation object can then be used to generate predictions at the
optimal value of
The cross validation object can then be used to generate predictions at the
optimal value of lambda:
enet_cv.predict(X')
In order to get double precision working without modifying Friedman's code,
some compiler trickery is required. The wrappers have been written such that
everything returned is expected to be a real*8 i.e. a double-precision
floating point number, and unfortunately the code is written in a way
Fortran is often written with simply real specified, letting the compiler
decide on the appropriate width. f2py assumes real are always 4 byte/
single precision, hence the manual change in the wrappers to real*8, but
that change requires the actual Fortran code to be compiled with 8-byte reals,
otherwise bad things will happen (the stack will be blown, program will hang
or segfault, etc.).
AFAIK, this package requires gfortran to build. g77 will not work as
it does not support -fdefault-real-8.
A build script has been provided in the glmnet/glmnet directory, so to build
the fortran extension:
cd glmnet/glmnet
source build.sh
Once the build is complete, you should have a _glmnet.so file in the glmnet
directory. Dropping into a python shell:
import _glmnet
Should work without error.
- Wrapper classes for the Poisson and Cox models.
Friedman's code in glmnet.f is released under the GPLv2, necessitating that
any code that uses it (including my wrapper, and anyone using my wrapper)
be released under the GPLv2 as well. See LICENSE for details.
That said, to the extent that they are useful in the absence of the GPL Fortran code (i.e. not very), the other portions may be used under the 3-clause BSD license.
- To Jerome Friedman for the fantastically fast and efficient Fortran code.
- To Pearu Peterson for writing
f2pyand answering my dumb questions. - To Dag Sverre Seljebotn for his help with
f2pywrangling. - To Kevin Jacobs for showing me his
wrapper
which helped me side-step some problems with the auto-generated
.pyf.
- J Friedman, T Hastie, R Tibshirani (2010). "Regularization Paths for Generalized Linear Models via Coordinate Descent".
- J Friedman, T Hastie, H Hofling, R Tibshirani (2007). "Pathwise Coordinate Optimization".