DeepCell is neural network library for single cell analysis, written in Python and built using TensorFlow and Keras.
DeepCell aids in biological analysis by automatically segmenting and classifying cells in optical microscopy images. The framework processes raw images and uniquely annotates each cell in the image. These annotations can be used to quantify a variety of cellular properties.
Read the documentation at deepcell.readthedocs.io.
For more information on deploying DeepCell in the cloud refer to the DeepCell Kiosk documentation.
| Raw Image | Tracked Image |
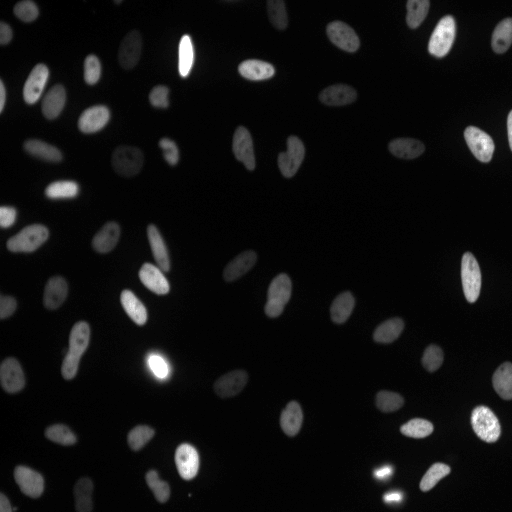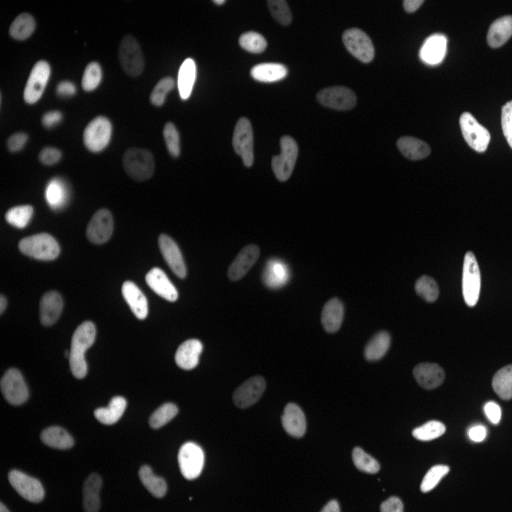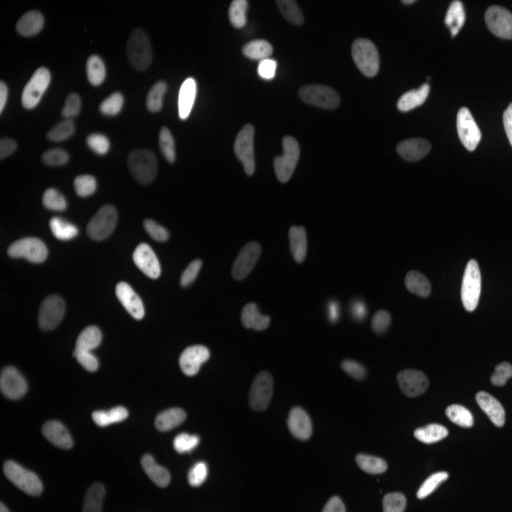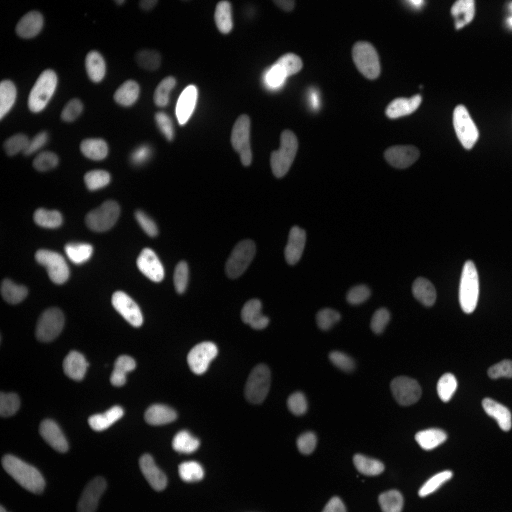
|

|
The fastest way to get started with DeepCell is to run the docker image:
nvidia-docker run -it --rm -p 8888:8888 vanvalenlab/deepcell-tf:0.4.0-gpuThis will start a Jupyter session, with several example notebooks detailing various training methods:
For examples of how to use the deepcell library, check out the following notebooks:
Together deepcell.datasets and deepcell.applications provide an accessible entrypoint to deep learning for biologists. The datasets module contains a variety of annotated datasets that can be used as training data. Additionally, the applications package initializes a set of models complete with the option to initialize with pre-trained weights.
DeepCell uses nvidia-docker and tensorflow to enable GPU processing. If using GCP, there are pre-built images which come with CUDA, docker, and nvidia-docker pre-installed. Otherwise, you will need to install docker, nvidia-docker, and CUDA separately.
git clone https://github.com/vanvalenlab/deepcell-tf.git
cd deepcell-tf
docker build --build-arg TF_VERSION=1.15.0-gpu -t $USER/deepcell-tf .# NV_GPU refers to the specific GPU to run DeepCell on, and is not required
NV_GPU='0' nvidia-docker run -it \
-p 8888:8888 \
$USER/deepcell-tf:0.4.0-gpuIt can also be helpful to mount the local copy of the repository and the scripts to speed up local development. However, if you are going to mount a local version of the repository, you must first run the docker image without the local repository mounted so that the c extensions can be compiled and then copied over to your local version.
# First run the docker image without mounting externally
NV_GPU='0' nvidia-docker run -it \
-p 8888:8888 \
$USER/deepcell-tf:latest
# Use ctrl-p, ctrl-q to exit the running docker image without shutting it down
# Then, get the container_id corresponding to the running image of deepcell
container_id=$(docker ps -q --filter ancestor="$USER/deepcell-tf")
# Copy the compiled c extensions into your local version of the codebase:
docker cp "$container_id:/usr/local/lib/python3.6/dist-packages/deepcell/utils/compute_overlap.cpython-36m-x86_64-linux-gnu.so" deepcell/utils/compute_overlap.cpython-36m-x86_64-linux-gnu.so
# close the running docker
docker kill $container_id
# you can now start the docker image with the code mounted for easy editing
NV_GPU='0' nvidia-docker run -it \
-p 8888:8888 \
-v $PWD/deepcell:/usr/local/lib/python3.6/dist-packages/deepcell/ \
-v $PWD/scripts:/notebooks \
-v /$PWD:/data \
$USER/deepcell-tf:0.4.0-gpu- The original DeepCell paper
- DeepCell 2.0: Automated cloud deployment of deep learning models for large-scale cellular image analysis
Copyright © 2016-2020 The Van Valen Lab at the California Institute of Technology (Caltech), with support from the Paul Allen Family Foundation, Google, & National Institutes of Health (NIH) under Grant U24CA224309-01. All rights reserved.
This software is licensed under a modified APACHE2. See LICENSE for full details.
All other trademarks referenced herein are the property of their respective owners.

