Project I worked on for a year and a half during my undergrad, coded in python. The underlying image manipulation libraries were c++ provided by VTK. The Project is used to create 3d printed moulds from a patients X-ray CT scans. These moulds lay on plastic mesh and deliver radiation therapy. I've published a paper on the project, which will be indexed by SPIE. https://github.com/Mark-William-Schumacher/Medical-3D-Printing/blob/master/Mark-Schumacher-Spie-Full-Submitted.pdf
You can also check out the Poster that I presented as the part of my Queens Computing Undergraduate Project.
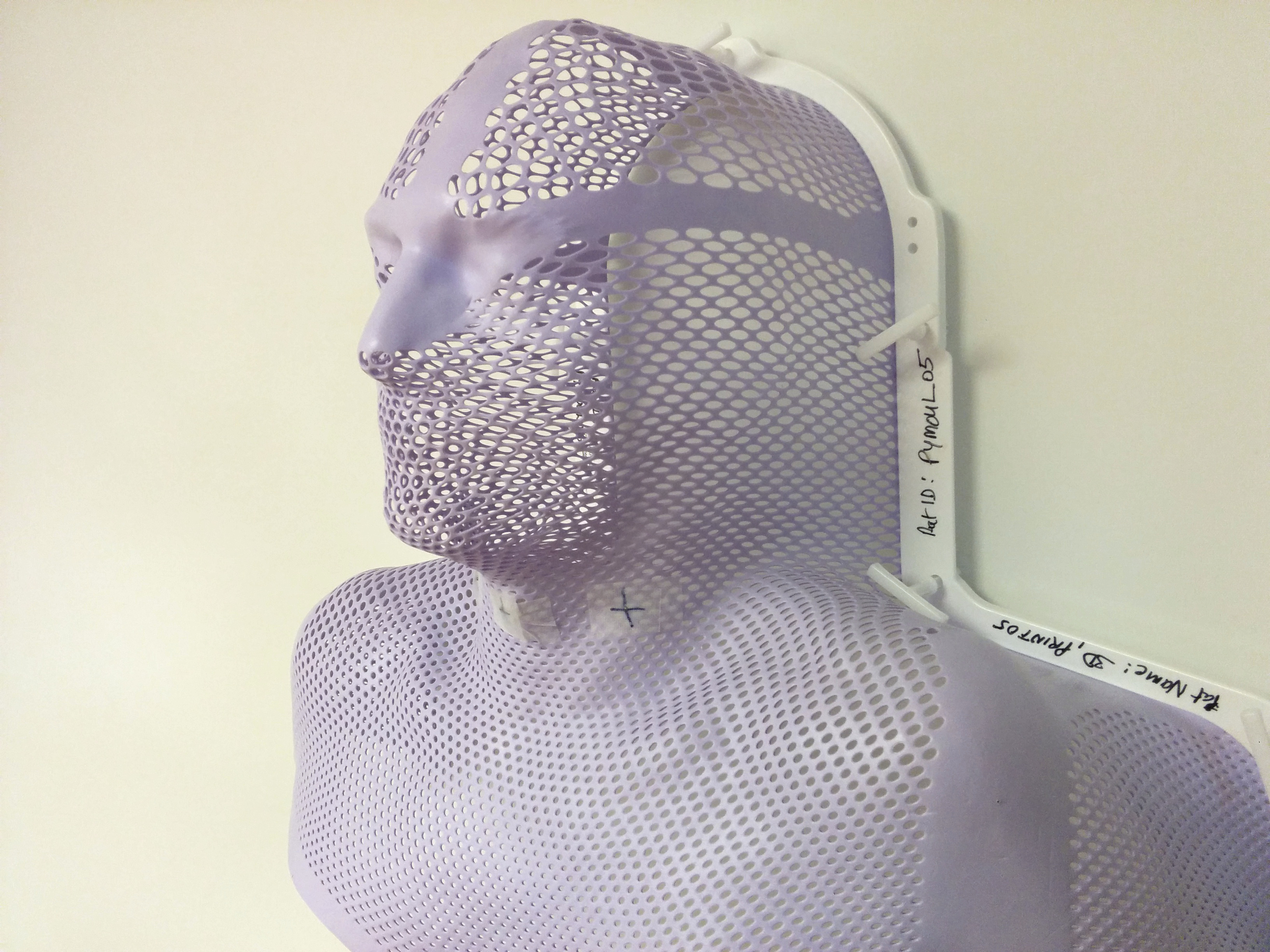
This is an example of the thermoplastic mesh we build the 3D moulds on, after taking any x-ray scan of this mesh, we can build a closed surface model and then generate a perfectly fitted mould to be placed on the mesh.
Most of this project was spent working in collaboration with my supervisor, but alot of the work done here was done on my own and I was responsible for both implementation and the direction of the project.
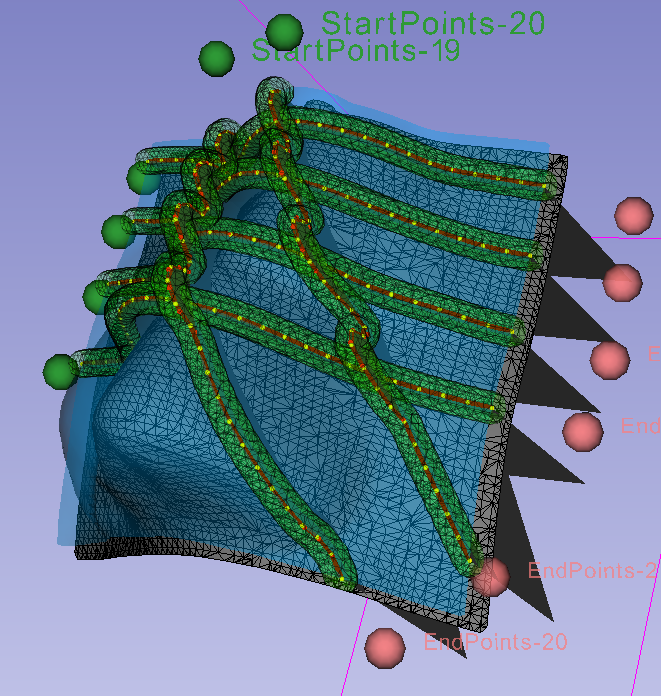
The algorythm works on the a closed surface model, which can be extracted from the x-ray image files using some semi-automated segmentation algorythms. The hospital technition would then define a region of interest and an array of startpoints and endpoints using Slicer's GUI interface.
This project can quiet a few interesting algorithms, and milestones. The first being the initial smoothing algorithm. The idea was that each of the paths which lay across the skin from start to end be smoothed in a way such that path never violated a minimum curvature. This is a crucial requirement because the catheters will be thread through 3D printout, and 'bending' them too much could stop the flow of radiation in portions of the mould and deliever lethal radiation to the patient. The path also cannot be smoothed using spline due to the fact that the this would cause the path to intersect with the patient face in some cases.
The smoothing algorythm, looks for the highest value concave point X and lifts the two adjacent neighbours to the point where X satisfies the minimum curvature requirement . When no concave points remain all remaining convex points which violate the minimum curvature can be solved with a single movement from the surface without cascading the violation.
The next challenge was performance, the project was taking to long to run (~20 minutes to generate a mould). After timing the functions the weak area of the process was combining the meshes. I opted to turn the meshes into binarized images to combine them, it took a lot of documentation reading in order to learn how to manipulate these large images for large scale boolean addition/shifting and transformations.
This is the final .STL image produced from running the algorythm described above.






.PNG)