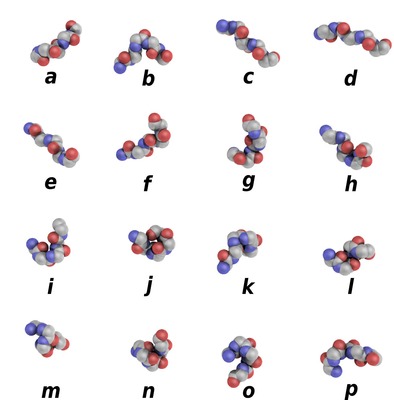
PBxplore is a suite of tools dedicated to Protein Block (PB) analysis. Protein Blocks are structural prototypes defined by de Brevern et al. The 3-dimensional local structure of a protein backbone can be modelized as an 1-dimensional sequence of PBs. In principle, any conformation of any amino acid could be represented by one of the sixteen available Protein Blocks (see Figure 1).
Figure 1. Schematic representation of the sixteen protein blocks, labeled from a to p (Creative commons 4.0 CC-BY).
PBxplore provides both a Python library and command-line tools. Basically, PBxplore can:
- assign PBs from either a PDB or either a molecular dynamics trajectory.
- use analysis tools to perform statistical analysis on PBs.
- use analysis tools to study protein flexibility and deformability.
For details, see the documentation at https://pbxplore.readthedocs.org/en/latest/.
PBxplore requires:
- Python 2.7 or Python 3.x (>= 3.3)
- the NumPy Python library,
Optionally, PBxplore can use:
- the MDAnalysis Python library (version >= 0.11) to read MD trajectories generated by Gromacs (.xtc files),
- the Matplotlib Python library to generate plots.
- WebLogo 3 to create logo from PB sequences.
Once dependencies installed, the most straightforward way is to use `pip`:
$ pip install pbxplorePBxplore can also be installed for the current user only:
$ pip install --user pbxploreAll documentation are hosted by Read The Docs and can be found here.
PBxplore is a research software and has been developped by:
- Pierre Poulain, DSIMB, Ets Poulain, Pointe-Noire, Congo
- Jonathan Barnoud, University of Groningen, Groningen, The Netherlands
- Hubert Santuz, DSIMB, Paris, France
- Alexandre G. de Brevern, DSIMB, Paris, France
If you want to report a bug, request a feature, use the GitHub issue system.
PBxplore is licensed under The MIT License.