- Run
preprocess.pyto isolate individual tumors from DICOM files with corresponding segmentations, resample to 1 mm pixel spacing, correct for low frequency intensity non-uniformity present in MRI data with the N4ITK bias field correction algorithm, and z-score normalize to reduce inter-scan bias. This saves individual images and masks as.nrrdfiles in addition to a dictionary linking anonymous patient identifiers to tumor-level imaging metrics. - Setup a
.csvfile identifying the input image and mask files to the pyradiomics extraction pipeline usingradiomic_setup.py. - Extract radiomic features using pyradiomics via the command line interface:
pyradiomics imagemaskfile.csv -o radiomicsresults.csv -f csv --jobs 16 --param params.yaml - Link clinical variables to imaging data with
link_clinical_imaging.py. - Compute various radiomic feature aggregation methods with
radiomic_aggregation.py. - Dimensionality reduction of radiomic features via minimum redundancy maximum relevance using
mRMRe_selection.R. - Load and format radiomic features selected via mRMRe with
selected_radiomic_loading.py. - Train models using
survival_models.py.
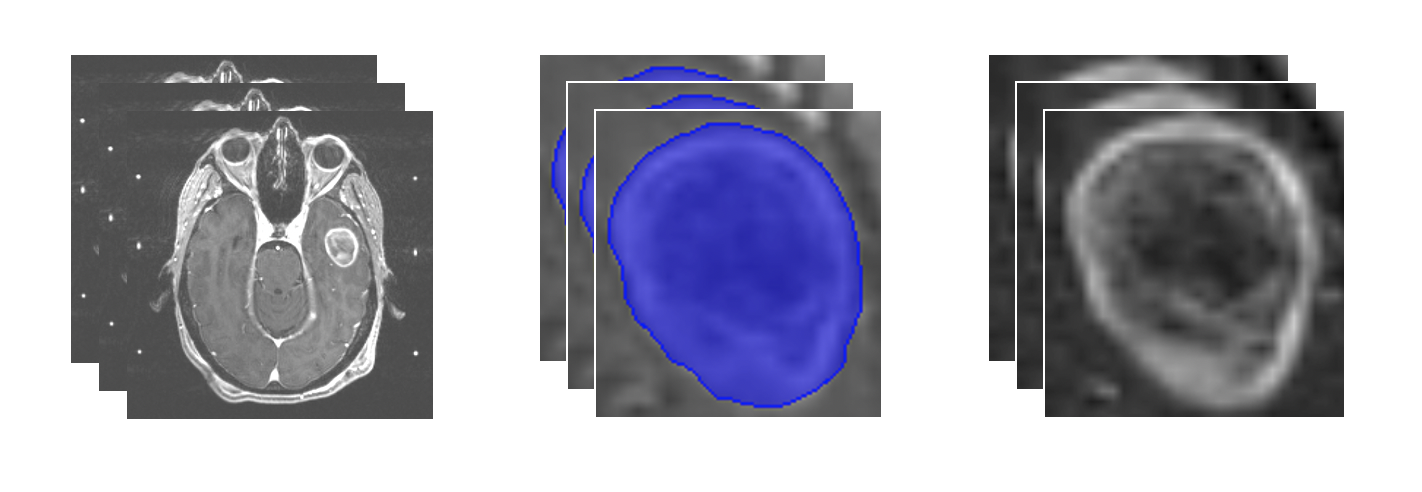
- Input: Slices of brain MRI scans loaded from DICOM files
- Identification of region of interest: Tumor segmentation
- Output: Extracted tumor of interest