Tools to produce, analyse and compare both simulated and recorded neuronal datasets
This repository is intended to capture the results of both performing analysis on the dynamics of c. elegans neurons that have been recorded, as well as analyzing the results of simulations of c. elegans neurons, and making the two comparable to each other. While our current simulations of c. elegans neurons still leave out many important details and tuning, there is a lot to be learned from even making coarse grained comparisons between real dynamics and dynamics that can come from imprecise models.
In a separate repo, progress was reported by @lukeczapla and @theideasmith in reproducing analysis via python code on existing data sets recorded from real c. elegans neurons:
Sample fluorescence heatmap for 107 neurons
 .
.
They wrote:
We also have some code to deal with the Kato data, which you can access at the link provided, in the folder wbdata. In that directory, if you
import transform.py as tf, you can access a dictionary containing all of Katos data withtf.wormData. The dictionary contains calcium imaging data for five worms and is keyed by the original matlab file names as they were sent by Kato himself :). Each worm's data's contains timevector ‘tv’, neural activity traces uncorrected (‘deltaFOverF') and corrected for bleaching (‘deltaFOverF_bc’) as well as derivatives (‘deltaFOverF_deriv'). The identity of neurons is in the same order in the cell array ‘NeuronIds'.
The same approach taken for recordings of real neurons can be applied to simulations. While the real neurons were reporting a fluorescence change over time, and the simulations are outputting membrane potential (voltage), at the level of gross dynamics, because those two variables are known to be related, they are comparable.
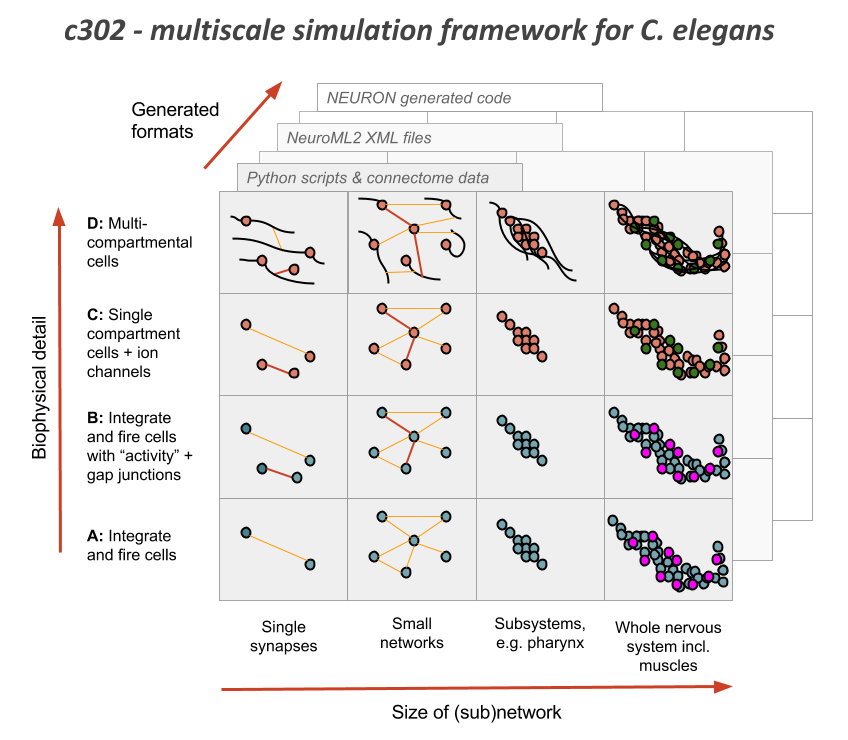
The OpenWorm project has been following a bottom-up approach to modeling the nervous system for some time. The c302 project has been a rallying point for this.
Using the libNeuroML library, combined with information about the name and connectivity of c. elegans neurons, we have built up a data structure that represents the nervous system of th ec. elegans, with details like the kinds of synapses, neurotransmitters, gap junction relationships it may have. Specifically, you can see how this is built in this script and this script.
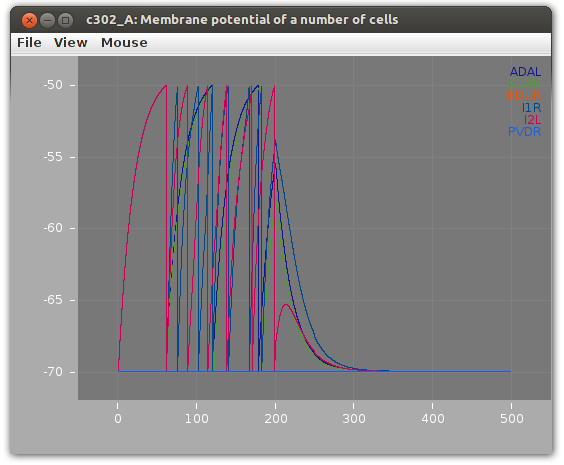
Then we can run the network as a simulation and see how the membrane potential might change over time.
The project of identifying and modeling the dynamics of all the ion channels has been the work of the ChannelWorm project. From there, the muscle model has been our rallying point for a highly detailed single cell model that would use our detailed ion channel models first. Beyond this, we have begun doing some work to improve the motor neurons and get the dynamics of the synapse between motor neurons and muscles corect. At each level, we have endeavored to constrain the model with data. Thus ion channel models are constrained by and tuned with I/V curve information mined from the literature. The same can begin to be done with the muscle model. While this is important infrastructure, there are still many gaps that are remaining.
With the advent of new experimental techniques to image hundreds of neurons at the same time, we now have another important kind of data to use to constrain the nervous system model. We can begin to explore what are the minimum dynamical conditions that are necessary to produce, in broad strokes, the same kinds of high-level dynamics. Even simplistic models of neurons hooked together with the c. elegans connectome will likely produce interesting investigations.
It is critical that the work of improving the nervous system model use all approaches, and result in a single unified model rather than a disconnected collection of models. While it is acceptable to have different versions of the models temporarily, eventually these must all converge on a single picture of the nervous system. To facilitate this, we have begun with modeling and computational infrastructure that is capable both of representing high-level and low-level aspects of the biophysics of neurons.
Next steps for this exciting endeavor will be described in the issues on this repo. As we collect scripts and code that are relevant to putting these pieces together, we will incorporate them here and make sure the community can execute them.