This dicoms anonymizer/pseudonymizer was made to make your life easier as a clinician sharing data.
The first goal is to remove the patient's name and replace by a pseudonym in both dicoms and a demographics csv file, but it also takes care of deleting other structures potentially containing identifiable information such as fields (eg, patient's phone number) and files (pdf, txt, csv, etc), via configurable blacklists.
WARNING: this project is not maintained anymore and is not fully compatible with Python 3. Please see alternative softwares if you want to ensure anonymization.
It runs under Python 2.7 (it was not tested nor developped with compatibility with Python 3 in mind, although it might work with some slight changes).
This is a generic dicoms pseudonymizer with an emphasis on:
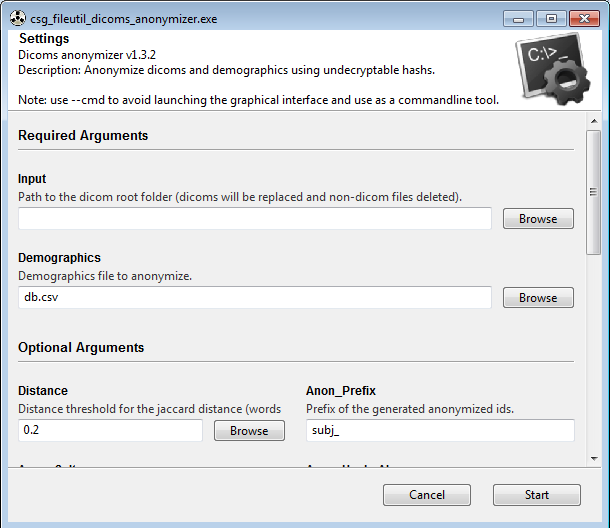
- Ease of use : simply point to the folder containing all your dicoms (folders or zip files) and to the demographics file (just ensure you have a "name" column), and click "Start"! Note that you can also use it in commandline, allowing batch scripting.
- Easy to anonymize demographics : a fuzzy matching algorithm will take care of small typos and of firstname/lastname reversal (works also if there are several first names or last names). At the end, several csv files will be generated: the anonymized demographics shortened to only the patients present in the dicoms (so that you can reuse a big demographics spreadsheet of all your subjects even to anonymize only a few subjects, the shortened anonymized demographics will only contain the pertinent subjects), the list of missing subjects that are in dicoms but not in demographics (so that your recipient knows directly what demographics were not available), and a mapping to convert from anonymized ids to original patient name (useful if your collaborator requests additional information about an anonymized patient, or just to check if there is an issue).
- Retaining potentially pertinent information in dicoms : instead of anonymizing all personal fields, this application anonymizes only the name (and strips out some other personal infos such as phone number, patient address, etc), but not all personal fields. Thus, anonymized dicoms will retain age, gender, date of scan, date of birth, etc. which can be then used as regression covariates or to cluster patients together. Additional fields to remove can easily be added as an argument.
- Reliability : a name with several typos or even a missing first name will still be anonymized correctly and linked to the correct demographics entry, by using fuzzy matching based on a normalized levenshstein distance for characters mistakes and a modified normalized words jaccard distance on words switching/missing. In any case, if a similar name cannot be found in the demographics, it will be replaced by a pseudonym and added to the mapping.
- Robustness : all other dicoms anonymizers look for the name only in the PatientName field. However, there are other hidden fields which can contain the patient's name, and this changes for each machine, each center, each parametrization of a scanner. This application will robustly find all fields containing the patient's name, and will try to remove it without touching the rest of the field (which can contain critical technical data), and it always checks that the name is cleared from the whole dicom before continuing. In case the patient name remains for whatever reason, the anonymizer will stop and show an error message so that you can take the appropriate steps.
- Deterministic one-way cryptography : pseudonyms are generated in a way that prevent any back tracing while still generating always the same pseudonym given the same patient's name. This is made possible by using a cryptographic hashing function with customizable salt: as long as you use a salt string specific to your organization, noone without this salt will be able to trace back the data without the mapping generated by this application, even if in possession of the registry of patients names.
To use this application, you must point to a folder where each subfolder is a patient dicom session. You can put multiple dicom sessions if it is for the same patient. This is because the script assumes one patient per folder (to look for name, else it would be too expensive to look at all dicoms files).
Optionally, you can provide a demographics file in csv format, to pseudonymize it along with dicoms, with the same pseudonyms. This csv file must include at least one column "name" where all the patients names are. Typos and words order do not matter, as there will be a fuzzy matching. However, note that this fuzzy matching might not work correctly if some names are too similar, or if there are too many typos or too many missing middle names. The fuzzy matching can be adjusted via the "dist_threshold" argument.
Finally, there are multiple additional options to refine the efficiency of the pseudonymization, for example there are blacklists for filetypes to remove that could contain identifiable informations (pdf, csv, txt, etc), that could be leftovers from the experimenter, as well as a blacklist for dicom fields to remove altogether (such as PatientPhoneNumber).
This app can work with a GUI or from commandline interchangeably, the same features and options will be available.
Another interesting feature is that you can build on top of an anonymized dataset, you can add new subjects. Indeed, each subject gets a unique id based on the name (by default) or by the order in the folder (by using the appropriate option), which makes it impossible to translate back to the original name from the id in any case.
The goal of this tool is to pseudonymize the data, not anonymize according to regulations. The difference is important: private fields will be kept (except if you specify a list of fields to delete), but patients names will be replaced by anonymous pseudonyms, rendering identification by name impossible (even when getting a hold of the subjects names, it is impossible without the mapping to trace back the demographics and dicom files to which names, thanks to the cryptographic salt). This is important for studies, where private fields contain important informations (scanner slice timing and parameters, patient's age and gender, etc).
If you want full anonymization (ie, removing/modifying all private fields), please have a look at dicom-anon, a Python dicoms anonymizer, with configurable specifications. See also pydicom example anonymizer and MATLAB's dicomanon command. Note however that this is not enough for full anonymization, as you will also need to deface (be warned this might make statistical preprocessing and analysis difficult).
To anonymize demographics for public release, you can have a look at the excellent ARX-Deidentifier, which supports various methods such as k-anonymity, l-diversity or differential privacy (this can be advantageously used as a post-processing step after csg-dicoms-anonymizer's ability to de-identify names robustly). To deface dicoms, Jorge's Defacer using nipy's quickshear algorithm and in pure Python 3 is an option to consider.
If you have only a handful of DICOMs you want to anonymize (eg, less than 20 subjects), you can do so manually using Dicom Browser.
This application is provided as-is with the hopes it can be useful to someone else. It was not cleaned up beforehand more than what was sufficient for running as a standalone GUI application and packaged as a minimal size binary.
With that in mind, the following should be done to make this application a proper software:
- CRITICAL: autodetect still images and offer option to delete them (they might contain images saved as pixels, currently the software will NOT autodetect nor remove them!)
- Make demographics file optional. It is already not really necessary in the code to anonymize dicoms, an empty csv with just the 'name' column should be enough. But it would be better if it was really optional and handled correctly.
- Make dicoms optional (enter an empty dicoms folder), so that a demographics file will still be anonymized.
- Allow to reuse an anonymized table (find similar names and assign the same hash).
- Refactor code: put functions above the code, delete the # [] useless comments coming from Jupyter export, move to functions some code snippets.
- Use requirements instead of integrating submodules in csg_fileutil_libs.
- Unit test with randomly generated dicoms.
- Make a dicoms reorganizer and deduplicator, using UUID (instance, study and machine?), good for sanity check too (so can organize per subject, scan and machine reliably, except if info was anonymized, can leave the option to dedup or not, can check by size too). See fields: Instance UUID, Study UUID, a third one (machine UUID?)
- Option to change all dates to relative date since another field chosen by user (be it birth, acquisition time, etc).
CSG Dicoms Anonymizer was initially made by Stephen Larroque <LRQ3000> for the Coma Science Group - GIGA Consciousness - CHU de Liege, Belgium. The application is licensed under MIT License.
If you found this software useful and use it for a publication, we would be very thankful if you could cite the software as follows, it's a free gesture for you but it tremendously help us in securing grants to further develop it:
Larroque, Stephen Karl. (2021, May 31). lrq3000/csg_dicoms_anonymizer: CSG Dicoms Anonymizer v1.4.2 (Version 1.4.2). Zenodo. https://doi.org/10.5281/zenodo.4885139