Copyright 2014-2017 Kiri Choi, J Kyle Medley, Matthias König, Kaylene Stocking, Caroline Cannistra, Michal Galdzicki, and Herbert Sauro
Parameter scan example
Tellurium is a python environment based on the Spyder2 IDE for building, simulating, and analysing models of biochemical networks, including gene regulatory networks, metabolic networks, and signaling pathways. It combines state-of-the-art scientific Python libraries, such as NumPy and SciPy, and includes special-purpose systems biology Python tools. Out of the box, Tellurium includes libRoadRunner, Antimony, PhraSED-ML, libSBML (via its Python bindings), and libSED-ML.
The Tellurium project is funded from the NIH/NIGMS (GM081070).
Tellurium can be installed via a front-end which includes all of its constituent packages, or via pip:
We recommend option 1. unless you are a developer and plan on integrating Tellurium with your current system.
- Download Tellurium notebook for Windows
- Double-click the installer to start the installation
- Follow the instructions
- Download Tellurium notebook for Mac OS X 10.10 or later
- Double-click the .dmg file to open a new window
- Drag the Tellurium icon to your Applications
- You can now launch Tellurium from e.g. Spotlight or your Applications folder directly
- TODO: Upload rpm
- Download the Debian package for Tellurium notebook
- Install the package using
dpkg -i Tellurium_2.0.0_amd64.deb - You should be able to launch Tellurium from your activities pane. If not, log out and in again or run
telluriumfrom the terminal.
- Download Tellurium IDE for Windows
- Double-click the installer to start the installation
- Follow the instructions
NOTE: Installation requires administrative rights. It is recommended to accept the default settings.
- Download Tellurium IDE for Mac OS X 10.10 or later
- Double-click the .dmg file to open a new window
- Double-click the Spyder icon
NOTE: On some older hardware we have noticed that this front-end sometimes fails. If this happens, we recommend using the Tellurium notebook front-end instead.
Binaries wheels for Tellurium are available via the pip command. To install Tellurium and the necessary packages on both Linux and OS X, type the following into a terminal:
pip install telluriumDetailed instructions for using the conda package manager are available here.
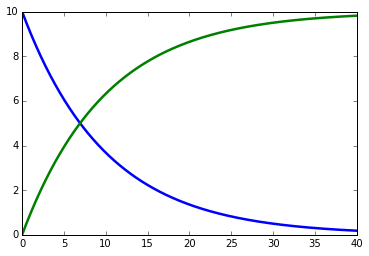
import tellurium as te
rr = te.loada('''
model example0
S1 -> S2; k1*S1
S1 = 10
S2 = 0
k1 = 0.1
end
''')
result = rr.simulate(0, 40, 500)
te.plotArray(result)
- General: http://tellurium.readthedocs.org/en/latest/
- API: http://tellurium.readthedocs.io/en/latest/_apidoc/tellurium.html
The Tellurium notebook supports Windows 10, Mac OS X 10.10+, Debian 8+, and Fedora 22+. The Spyder IDE installers are tested with Windows 7+ and Mac OS X 10.9+. Some older Macs cannot run Spyder, regardless of whether the operating system is up-to-date. Pip packages are tested on Fedora 22, Debian 8, Ubuntu 14.04, and Mac OS X 10.10.
The Tellurium PyPI packages support 64-bit Python versions 2.7, 3.4, 3.5, and 3.6 for Windows, Mac, and Linux. The notebook viewer comes with Python 3.6 (64-bit) and the IDE comes with Python 2.7 (32-bit).
The source code for the Tellurium Python package is hosted at https://github.com/sys-bio/tellurium and is licensed under the Apache License, Version 2.0. Tellurium uses third-party dependencies which may be licensed under different terms. Consult the documentation for the respective third-party packages for more details.
TELLURIUM AND ALL SOFTWARE BUNDLED WITH TELLURIUM (HEREAFTER "THE SOFTWARE") IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDERS OR ANYONE DISTRIBUTING THE SOFTWARE BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THE SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.