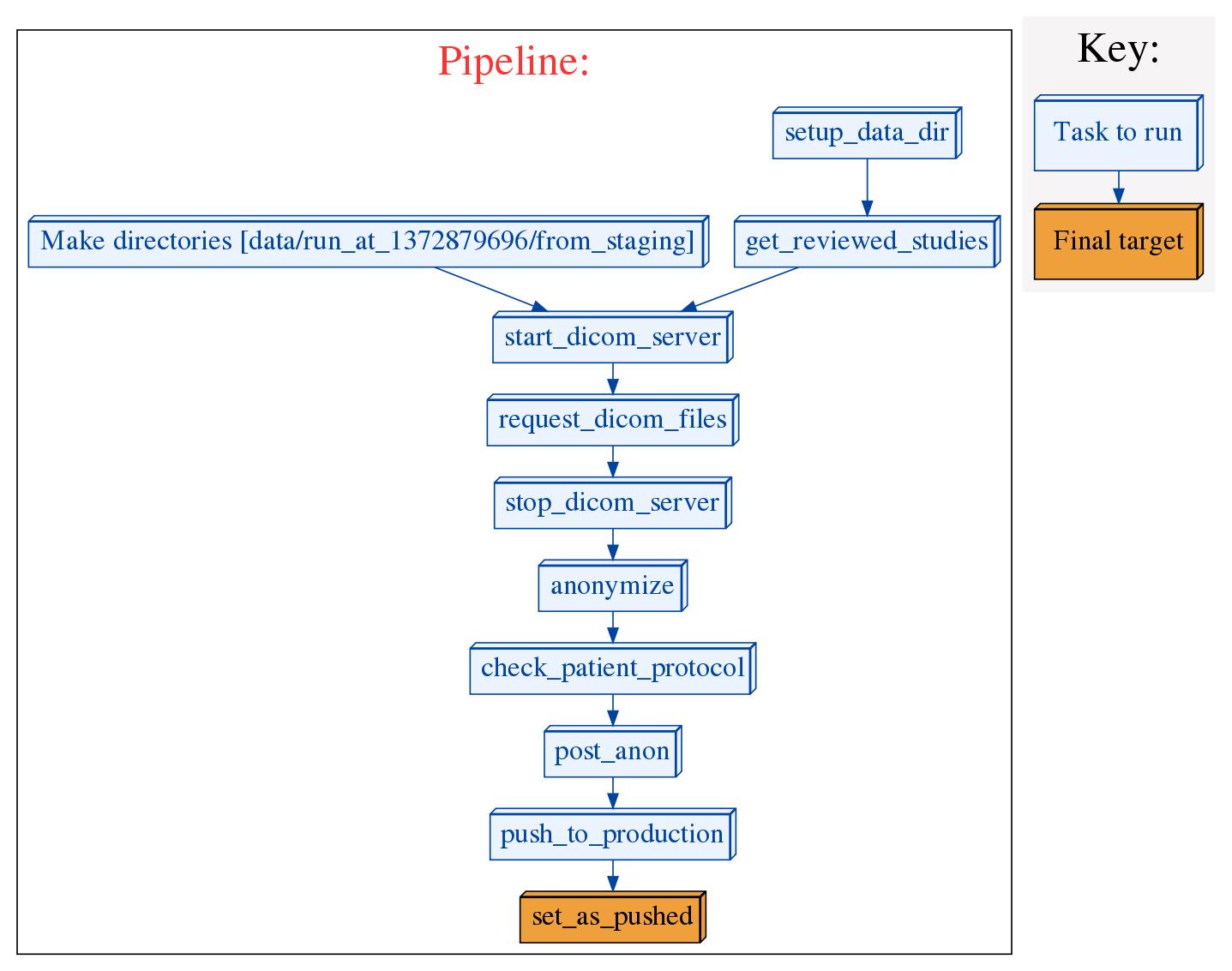
This is a software pipeline meant to perform the following steps on DICOM studies after they have been reviewed with django-dicom-review
- Pull the studies from a staging (identified) PACS
- Anonymize the studies
- Integrate with existing research databases/other post anonymization task
- Push the studies to a research PACS
The pipeline records information about each step using log files in a time stamped directory (more details to follow). It is capable of restarting a run from a failed step.
This pipeline is currently using a very new DICOM anonymizer (located at http://github.com/cbmi/dicom-anon). Please use with caution and report any issues.
This pipeline assumes you have two image archives (PACS), one where identified images are stored (staging) and one where the de-identified images will be stored (production). It is assumed that you are running the django-dicom-review application so that it serves up to reviewers images from the identified staging archive, and this pipeline will be pushing to the production staging archive.
-
A recent version of Java.
-
The dcm4che2 toolkit must be downloaded and the bin directory from the toolkit must be on the system path.
-
Python version 2.7.1 (2.6 may work, but has not been tested). Pip must also be installed. It is highly recommended that the pipeline be installed in a clean virtualenv.
-
git
-
VERY IMPORTANT The value you are using for "LOCAL_AE", which by default is "DCMQR" (but can be changed in your local_settings.py file, see below) must be setup on your staging PACS to point to the IP address of the machine the pipeline is running on. If you are using DCM4CHEE this can be done via the AE Management tool on the web console. If this is not set properly you could send identified DICOM data to the wrong machine.
Run the following commands to install the pipeline. If you are using a virtualenv, you may want to clone it into that directory.
git clone https://github.com/cbmi/dicom-pipeline.git dicom_pipeline
cd dicom_pipeline
git submodule update --init
pip install -U -r requirements.txt
Once this is done, you will need to place a valid local_settings.py in your root directory.
This settings file is used by both the Django ORM to read and write to the models that represent your studies and the DICOM utilities that receive and push the actual files to the PACS.
In the root directory of the pipeline repository you will find a local_settings.sample.py. Rename this file local_settings.py and fill out as appropriate. The comments within the file should explain what each value represents. Again, the "LOCAL_AE" value in this file must be setup properly on your staging PACS as described in the pre-requisites section.
If you have an existing identity.db (which is used by the anonymizer), it must be in the pipeline root directory. Otherwise it will create a new one, and it will not be able to take advantage of any previous mappings it made. For example, if you use the same identity.db file, and the anonymizer previously mapped identified Study UID 1 to anonymized Study UID 0, it will continue to use that same mapping in the current round of anonymizations. If you do not have the identity.db file, STUDY UID 1 will be re-assigned a new anonymized Study UID if encountered.
The pipeline.py file has a few options, including the ability to limit the number of studies processed (-m), restarting the last pipeline attempt instead of starting a new one (-r), and running a practice run that makes no permanent changes (-p). Use the --help option to see details on all options.
From the root directory of the pipeline git repository, run the following command
python pipeline.py
When the pipeline is run, it will create a directory called data in the current working directory (if it does not already exist), and within that directory, it will create a directory called run_at_<seconds_since_epoch>. This directory will be the working directory for that particular run of the pipeline. In this directory, as the pipeline progresses it will created files and directories that make up a trail of what the pipeline did. The following directories will be created:
- from_staging - this will contain all the identified DICOM files pulled from your staging PACS
- quarantine - this will contain files that the anonymizer deems as likely to contain PHI and that will not be pushed to production
- to_production - this directory will contain the anonymized files that were pushed to production
The following files will also be created:
-
overview.text - This file will contain a summary of how the run went. Here is a sample:
Starting at 2013-05-03 08:00 100 valid reviewed studies. Please review comments.txt Received 23948 files containing 100 studies 1 studies marked as having a protocol series, 1 studies found with protocol series during anonymization. 1 studies marked as having a protocol series but not found, see 'missing_protocol_studies.txt'. 22755 files containing 96 studies were successfully hooked up to an encounter. Push completed at 2013-05-03 15:15 -
comments.txt - This file just contains all the studyuids and associated reviewer comments for each. It is primarily to quickly review anything reviewers may have put in comments.
-
studies_to_retrieve.txt - This contains all the studyuids the pipeline tried to retrieve from staging.
-
pull_output.txt - Output from the DICOM staging pull process.
-
anonymize_output.txt - Output from the anonymizer process
-
post_anon_output.txt - Output from the post anonymization process. You can control the code that is executed here, see the next section for details.
-
push_output.txt - Output from the push to production process
-
found_protocol_studies.txt - This is a list of all the studies found to have a series called Patient Protocol (which in our experience is never something you want pushed to production)
-
missing_protocol_studies.txt - If the reviewer marked a study as containing a protocol study in the review app, but it was not found during anonymization, this file will list the study uid for further review.
-
reviewed_protocol_studies.txt - A list of all the studies marked as containing a protocol study by the reviewers.
The most institution/project specific step in the pipeline is the post_anonymize step. For our internal implementation, and by default, this will execute the script called load_studies.py which uses the identity.db file created and maintained by the anonymizer to hook up the now de-identified studies to existing patients in a research database. It is likely that other projects will need to perform a similar task, but unlikely that the process will be the same as database schemas in existing systems will vary. The pipeline includes a hook to enable overriding the code that gets executed at this step.
-
Create a file called custom_hooks.py in the root pipeline directory.
At the top of the file do the following imports
from hooks import registry
-
Define a function in the file that you want to be executed after the DICOM files have been anonymized. It should have the following signature (the function name does not matter):
def custom_hook(run_dir, overview, practice): """ run_dir - String containing the path of the working directory for this pipeline run. Anonymized files will be in a directory within that directory called 'to_production'. use os.path.sep.join([run_dir, 'to_production']) to get the full path. overview - Stream object for printing updates that will be placed in the overview.text for the pipeline run practice - Boolean, whether or not this a practice run. Returns - Should return a string that will be placed in a file called post_anon_output.text which can be audited later. This is for a higher level of detail than what is written to the overview stream. """ return "post anon complete"
-
At the bottom of the file, register your function with the system:
registry.register(custom_hook, name = 'my_custom_hook')
-
Finally, in your
local_settings.pyfile, setPOST_ANON_HOOKequal to the name you used in the above register command:POST_ANON_HOOK = 'my_custom_hook'
The process of linking de-identified studies to existing de-identified patients will likely require that you to trace from the de-identified studyuids and other attributes to the identified ones. To do this, you can query the sqlite database created by the pipeline. This creates tables for some of the DICOM attributes that were cleaned with an original and cleaned field for each value that was changed. You can examine the database using the sqlite client, but some of the tables it creates are patient, accession_no, and studyuid. For example, if in your post_anon_hook, you were looking at a de-identified DICOM file and you needed to find out what its original accession_no was, you could issue the following SQL query to the identity.db.
select original from accession_no where cleaned = '<cleaned_accession_no>';
Some DICOM values can potentially contain PHI, but completely stripping them can reduce the utility of the studies for research. This is especially true for Study and Series descriptions because it makes it difficult to tell what the study contains. To accommodate this, the anonymizer allows you to enforce white lists on specific DICOM attributes. If the value matches one on the white list, it will be left, if not, it will be stripped. The dictionary is stored in a file called dicom_limited_vocabulary.json. It is a dictionary where the keys are DICOM attributes in the format "0000,0000", and the values are lists of strings that are allowed for that value. By default, it is an empty dictionary, so nothing will be enforced. Below is an example that would allow a couple values for Study and Series description.
{
"0008,1030": [
"CT CHEST W/CONTRAST",
"NECK STUDY"
],
"0008,103E": [
"3D HEAD BONE",
"ST HEAD"
]
}The clean_data.py script takes a date as a command line argument and deletes the run_at directories in your data directory that were created earlier than that day. This can be used to save space by cleaning out old DICOM files that are no longer needed. The text log files will remain.
There are various scripts in the dicom-tools repository (which is checked out as a submodule of this repo). See the README in that repository. The two most directly useful to DICOM anonymization are:
- filter_dicom.py - Selectively move certain DICOM files by specifying study or series id to a different directory. This can be used to remove certain files before pushing to production.
- print_summary.py - Print out all the unique study/series descriptions and patient names for all DICOM files in a directory.