MLHOUSE is a neural network that is trained to predict the high-resolution Rosetta energy of a protein with low-resolution modeling. In short, each residue in the protein is represented by a single sphere as opposed to having a sphere for every atom in the residue.
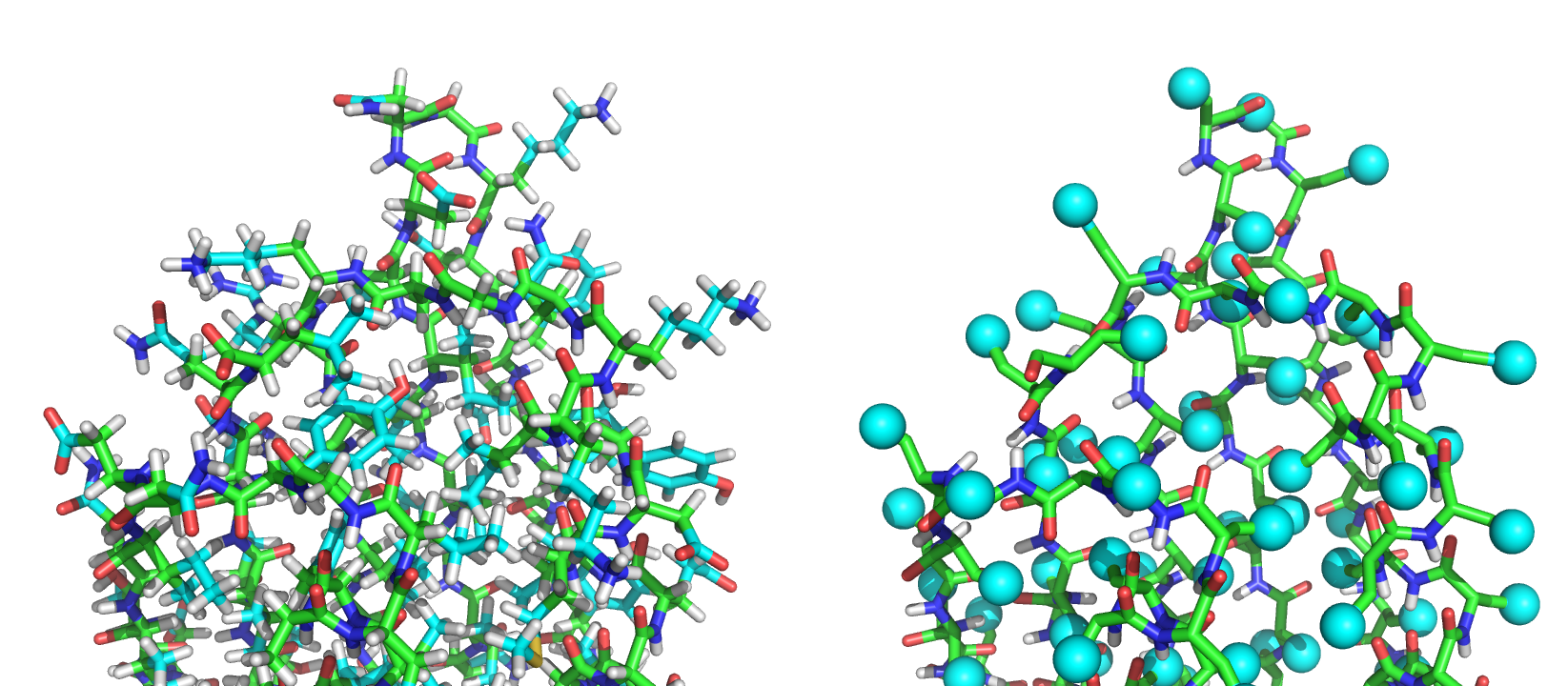
The PDB 1QYS in the (L) full atom representation, and (R) the centroid representation
MLHOUSE operates by predicting the energy of each surface residue iteratively. To get the prediction for one residue, MLHOUSE casts rays in all directions from the center of that residue's centroid sphere. MLHOUSE performs a simple ray-tracing algorithm to detect the first centroid sphere (belonging to a different residue) each ray hits. Instead of deflecting like a normal ray-tracing algorithm, this ray stops and returns properties about the sphere it hits (outlines in the nexted section).
(A) In centroid (or "low resolution") mode, each residue is represented by one sphere. This picture shows 4 residues.
(B) To get the energy for the middle residue, MLHOUSE casts rays in all directions. Each ray stops when it hits another sphere.
(C) Working clockwise from the 12:00 position, this is what the "Distance vs Angle" plot looks like. You can see the distance decrease when it hits a sphere, then return to the maximum value (rays don't travel more than 16 Å).
(D) We see a much clearer picture of the spheres when we increase the sampling resolution from 45 degrees to 1 degree.
The cartoon above only casts rays in 2D space, resulting in a 1D vector (subfigure D). MLHOUSE operates in 3D space, so the rays create a 2D vector that can be interpretted as an image (see below). This image is then fed into a deep neural network that is trained to predict two properties: (1) the Rosetta energy of the residue after performing fixed-backbone rotamer substitution (known as "packing", can incorporate design) and (2) the binding energy after packing is performed in the bound state.
The following pictures were generated by sampling residue 50 (using Rosetta/absolute numbering, not PDB numbering) of PDB code 3U3B.
Darker objects represent shorter distances.
Black represents being close to the backbone of the sphere, white represents being on the sidechain end of the sphere.
This one is complicated, but it is meant to portray the relationship between the ray and the center of the intersecting sphere. Black represents a smaller value of Thc (in the picture below) and white is a larger value.
1 if the interesecting sphere is part of the same chain as the source sphere, otherwise -1. -1 is also returned if the ray does not interest with a sphere, however that case is treated differently in this picture for the sake of visual clarity.
The white spheres in the picture are part of the same chain as the source sphere, the gray spheres are part of a different chain, and (for the sake of this image) the background is black.
This image is generated for each of the 20 amino acids. Residues that can adopt that amino acid identity are colored white, even if they have a different amino acid identity to start out. A residue position that can adopt all 20 amino acids will be white in every picture.
The pictures shown above are naturally distorted so that the north and south poles are stretched wide. Here is a less distorted representation of the distance map:
The pictures above are shown at a higher resolution than MLHOUSE actually samples. They were generated by sampling both axes with 1 degree resolution (360 x 180), but the algorithm uses 9 degree resolution (40 x 20) as shown below.
Rosetta's Full Atom Representation vs Centroid Representation