=========
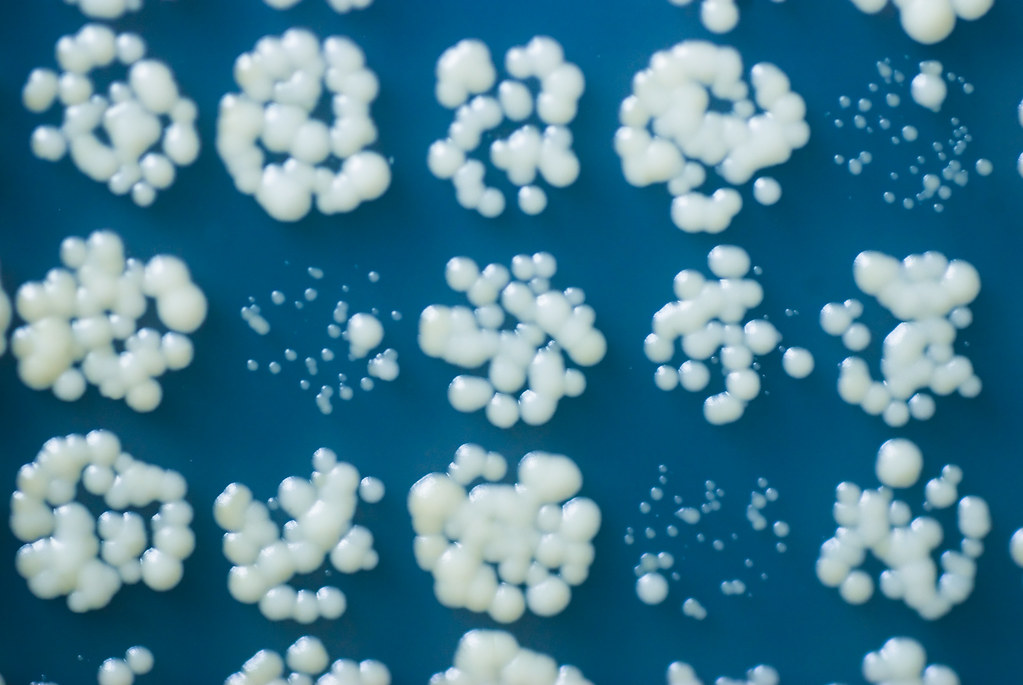
Image analysis software for estimating cell density in arrayed microbial cultures growing on solid agar
Colonyzer is currently undergoing major redevelopment
- Python 3 compatible - installation via pip more straightforward
- New command-line interface - separate analysis scripts merged into improved command-line structure
- Improved documentation - access hints about options directly on the command line
- Easier grid location - only specify gridded array format, Colonyzer.txt calibration files should not be necessary for most images
- Improved grid location algorithms - estimate grid width by autocorrelation analysis, grid location and rotation by gradient-based optimisation (with optional global search)
- Improved spot location algorithm - recursive estimation of centre of mass of each spot, subject to no increase in signal along tile edge, refines grid location estimates for irregular spots
- Alternative segmentation algorithm - detects cells by first detecting culture edges and infilling: even higher sensitivity at very low signal levels
- Improved lighting correction - generate pseudo-empty plate by infilling segmented pixels using a Gaussian Random Markov Field update
- Test datasets - suite of problematic images for checking performance of scripts and algorithms
Website describing current, stable release: http://research.ncl.ac.uk/colonyzer/
Open access manuscript describing Colonyzer algorithms: http://dx.doi.org/10.1186/1471-2105-11-287
Open access video and manuscript demonstrating the use of Colonyzer within a Quantitative Fitness Analysis workflow http://www.jove.com/video/4018