df_org = df.copy()
df = df.dropna()
df['Customer_Location'] = df['Customer_Location'].astype("category").cat.codes
X = df.drop(['Email_Status'], axis=1)
Y = df['Email_Status']
## Oversampling Minority classes
from imblearn.over_sampling import SMOTE
df_m = df.copy()
df_m = df_m[(df_m.Email_Status == 1) | (df_m.Email_Status == 2)]
X_m = df_m.drop(['Email_Status'], axis=1)
Y_m = df_m['Email_Status']
X = X.loc[Y[Y==0].index]
Y = Y[Y==0]
sm = SMOTE(random_state=np.random.randint(0, 100))
X_os_m , Y_os_m = sm .fit_resample(X_m, Y_m)
X_os = pd.concat([X, pd.DataFrame(X_os_m, columns= X.columns)], axis=0)
Y_os = pd.concat([Y, pd.Series(Y_os_m)], axis=0)
X_train, X_test, y_train, y_test = train_test_split(X_os, Y_os,
test_size=0.3,
random_state=71, stratify=Y_os)
def hyperopt_train_test(params):
t = params['type']
del params['type']
if t == 'naive_bayes':
clf = BernoulliNB(**params)
elif t == 'svm':
clf = SVC(**params)
# label: match value in binary
y = data['match_x']
print(y)
a = np.where(x.values >= np.finfo(np.float64).max)
print(a)
# convert pandas dataframe to numpy matrix
x = x.values.astype('float64')
y = y.values.astype('float64')
# ======================== SMOTE Oversampling ========================
print("[INFO] SMOTE Oversampling")
print("Original Dataset: ", binary_counter(y)) # count of +ve and -ve labels
sm = SMOTE(random_state=209)
x, y = sm.fit_sample(x, y)
print("SMOTE Resampled Dataset: ", binary_counter(y))
x_train, x_test, y_train, y_test = train_test_split(x,
y,
test_size=0.1,
random_state=42)
norm = True
# ------ Normalize data ------
if norm:
x_train = normalize(x_train)
x_test = normalize(x_test)
import numpy as np
from sklearn.preprocessing import StandardScaler
import random
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import cross_validate
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.externals import joblib
data = pd.read_csv("indian_liver_patient.csv")
data = data.fillna(method="ffill")
data.Gender = data.Gender.map({"Female": 1, "Male": 0})
data["Dataset"] = data["Dataset"].map({1: 0, 2: 1})
np.random.shuffle(data.values)
print(data.columns)
target = data["Dataset"]
source = data.drop(["Dataset"], axis=1)
sm = SMOTE()
sc = StandardScaler()
lr = LogisticRegression()
source = sc.fit_transform(source)
X_train, X_test, y_train, y_test = train_test_split(source,
target,
test_size=0.01)
X_train, y_train = sm.fit_sample(X_train, y_train)
cv = cross_validate(lr, X_train, y_train, cv=10)
print(cv)
joblib.dump(lr, "model4")
def class_imbalance(X_data, y_data):
# creating an instance
sm = SMOTE(random_state=27)
# applying it to the data
X_train_smote, y_train_smote = sm.fit_sample(X_data, y_data)
return X_train_smote, y_train_smote
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import classification_report, f1_score, precision_score, recall_score
from sklearn.ensemble import RandomForestClassifier
from collections import Counter
from imblearn.ensemble import EasyEnsemble
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import cross_val_score
RANDOM_STATE = 500
bottlenecks = pd.read_csv('./bottlenecks_train.csv')
targets = pd.read_csv('./target.csv')
daibao = targets['daibao']
X_test = pd.read_csv('./bottlenecks.csv')
smo = SMOTE(random_state=RANDOM_STATE)
X_train, y_db_train = smo.fit_sample(bottlenecks, daibao)
ss = StandardScaler()
X_train = ss.fit_transform(X_train)
X_test = ss.transform(X_test)
# print(X_train.shape)
t1 = time.time()
clf = RandomForestClassifier(n_estimators=50,
max_depth=20,
random_state=RANDOM_STATE).fit(
X_train, y_db_train)
pro = clf.predict_proba(X_test)
# Seperate inputs and outputs
labels = df[34]
inputs = df.drop([34], axis=1)
# Seperate data into a training/test set
input_train, input_test, output_train, output_test = train_test_split(
inputs, labels, test_size=0.33, random_state=42)
# %%
print("Before OverSampling, counts of label '1': {}".format(
sum(output_train == 1)))
print("Before OverSampling, counts of label '0': {} \n".format(
sum(output_train == 0)))
sm = SMOTE(random_state=27, sampling_strategy=1.0)
input_train_res, output_train_res = sm.fit_sample(input_train,
output_train.ravel())
print("After OverSampling, counts of label '1': {}".format(
sum(output_train_res == 1)))
print("After OverSampling, counts of label '0': {}".format(
sum(output_train_res == 0)))
#Dimentioanlity reduction
from sklearn.decomposition import PCA
pca = PCA()
input_train = pca.fit_transform(input_train)
input_test = pca.transform(input_test)
total = sum(pca.explained_variance_)
knn_with_gridSearchCV(
X,
y,
K=K,
T=T,
R1=R1,
R2=R2,
test_ratio=0.2,
filename='knn_with_gridSearchCV_with_full_dataset.png')
# Remove any label with members < 2
data = data[data.groupby('quality').quality.transform('count') >= 2].copy()
## Fix imbalance dataset
[X, y] = separate_data_from_label(data)
X_res, y_res = SMOTE(k_neighbors=3, random_state=0).fit_resample(X, y)
print("Original data shape: X.shape = ", X.shape, "y.shape = ", y.shape)
print("New data shape: X.shape = ", X_res.shape, "y.shape = ", y_res.shape)
## KNN ##
knn(X_res, y_res, K, filename='knn_fix_imbalance_dataset.png')
print()
## KNN WITH CROSS VALIDATION ##
knn_with_cross_validation(X_res, y_res, K, T)
## GridsearchCV ##
knn_with_gridSearchCV(
X_res,
y_res,
K=K,
#opt=optimizers.Adam(lr=1e-4)
'''
labels=['Non-Hazardous','Hazardous']
NNperformance(init_mode,act,opt,n_top_features,epochs,batch_size,labels,X_train_sfs_scaled, y_train,X_test_sfs_scaled, y_test)
from imblearn.over_sampling import SMOTE,RandomOverSampler,BorderlineSMOTE
from imblearn.under_sampling import NearMiss,RandomUnderSampler
smt = SMOTE()
nr = NearMiss()
bsmt=BorderlineSMOTE(random_state=42)
ros=RandomOverSampler(random_state=42)
rus=RandomUnderSampler(random_state=42)
X_train_bal, y_train_bal = bsmt.fit_sample(X_train_sfs_scaled, y_train)
print(np.bincount(y_train_bal))
NNperformance(init_mode,act,opt,n_top_features,epochs,batch_size,labels,X_train_bal, y_train_bal,X_test_sfs_scaled, y_test)
#Plot decision region
def plot_classification(model,X_t,y_t):
clf=model
pca = PCA(n_components = 2)
X_t2 = pca.fit_transform(X_t)
params['data_path'] = r'F:\PythonProject\MobileNet\BearingProject\BearingData\feature\traindata_N15_M07_F10_fea2.csv'
params['save_path'] = r'F:\PythonProject\MobileNet\BearingProject\BearingData\smote_feature\traindata_N15_M07_F10_fea2.csv'
# X为特征,y为对应的标签
train = pd.DataFrame(pd.read_csv(params['data_path']))
y = train['label']
X = train.loc[:, :'ratio_cD1']
from collections import Counter
# 查看所生成的样本类别分布,0和1样本比例9比1,属于类别不平衡数据
print(Counter(y))
# Counter({0: 900, 1: 100})
# 使用imlbearn库中上采样方法中的SMOTE接口
from imblearn.over_sampling import SMOTE
# 定义SMOTE模型,random_state相当于随机数种子的作用
smo = SMOTE(random_state=42)
X_smo, y_smo = smo.fit_sample(X, y)
y_smo=y_smo.reshape(-1,1)
#X_smo1=pd.DataFrame(X_smo)
#y_smo1=pd.DataFrame(y_smo)
#print(np.size(X_smo))
#print(np.size(y_smo))
smo_train = np.concatenate((X_smo,y_smo),axis=1)
# 查看经过SMOTE之后的数据分布
#print(Counter(y_smo))
# Counter({0: 900, 1: 900})
#col_lab = ['time_mean','time_std','time_max','time_min','time_rms','time_ptp','time_median','time_iqr','time_pr','time_sknew','time_kurtosis','time_var','time_amp','time_smr','time_wavefactor','time_peakfactor','time_pulse','time_margin',
# 'freq_mean','freq_std','freq_max','freq_min','freq_rms','freq_median','freq_iqr','freq_pr','freq_f2','freq_f3','freq_f4','freq_f5','freq_f6','freq_f7','freq_f8',
# 'ener_cA5','ener_cD1','ener_cD2','ener_cD3','ener_cD4','ener_cD5','ratio_cA5','ratio_cD1','ratio_cD2','ratio_cD3','ratio_cD4','ratio_cD5','label']
col_lab = ['time_mean','time_median','freq_mean','freq_std','freq_median','freq_f2','freq_f5','freq_f6','freq_f7','freq_f8','ener_cD1','ratio_cD1','label']
modelsList = []
for idx in range(opt.num_models):
print('Ensemble Model', idx)
print('\tResampling model training data')
trainX, validateX, trainY, validateY = train_test_split(trainX,
trainY,
test_size=0.2)
#smt = SMOTETomek(sampling_strategy='auto')
#smt = RandomUnderSampler(sampling_strategy='auto')
#smt = TomekLinks(sampling_strategy='auto')
#smt = ClusterCentroids(sampling_strategy='auto')
#smt = EditedNearestNeighbours(sampling_strategy='auto', n_neighbors=9)
smt = SMOTE(sampling_strategy='auto', k_neighbors=5)
#smt = SMOTEENN(sampling_strategy='auto', smote=None, enn=None)
X_smt, y_smt = smt.fit_resample(trainX, trainY)
#X_smt, y_smt = trainX, trainY
train_data = []
for i in range(len(X_smt)):
train_data.append([X_smt[i], y_smt[i]])
test_data = []
for i in range(len(testX)):
test_data.append([testX[i], testY[i]])
training_data_loader = DataLoader(dataset=train_data,
num_workers=opt.threads,
def preproc_data(X_train,
y_train,
resample_training,
n_poly,
scale_x,
scale_cols,
X_test=None,
fsel=False,
combine_cols=None):
# give feat lists (dictionary of lists) to combine cols if you want to combine things
# Optionally add in interactions/n order polynomial features
# ****(right now this does to all -- need to fix!!!)***
if n_poly:
poly_feat = PolynomialFeatures(degree=n_poly, include_bias=False)
X_train = poly_feat.fit_transform(X_train)
if X_test is not None:
X_test = poly_feat.transform(X_test)
# Optionally scale features
if scale_x:
print('Scaling X...')
robust_scaler = StandardScaler()
# for numeric cols, fit model to training data
X_train.loc[:, scale_cols] = robust_scaler.fit_transform(
X_train.loc[:, scale_cols])
# now update test if necessary
if X_test is not None:
X_test.loc[:, scale_cols] = robust_scaler.transform(
X_test.loc[:, scale_cols])
# optionally combine similar features
if combine_cols is not None:
print(X_train.head())
for new_name in combine_cols.keys():
print('Creating ', new_name)
feat_list = combine_cols[new_name]
print(feat_list)
X_train = combine_feats(X_train, feat_list, new_name)
if X_test is not None:
X_test = combine_feats(X_test, feat_list, new_name)
# optionally upsample training
col_names = X_train.columns
if resample_training == 'over':
print('Upsampling with SMOTE...')
sm = SMOTE() # Synthetic Minority Over-sampling Technique
X_train, y_train = sm.fit_sample(X_train.as_matrix(),
y_train.as_matrix())
X_train = pd.DataFrame(X_train,
columns=col_names) # convert back to pandas
elif resample_training == 'under':
print('Undersampling randomly...')
rus = RandomUnderSampler()
X_train, y_train = rus.fit_sample(X_train.as_matrix(),
y_train.as_matrix())
X_train = pd.DataFrame(X_train,
columns=col_names) # convert back to pandas
if fsel:
selector = SelectKBest(f_classif, k=25)
selector.fit(X_train, y_train)
# Get idxs of columns to keep
idxs_selected = selector.get_support(indices=True)
# overwrite existing dataframe with only desired columns
X_train = X_train.iloc[:, idxs_selected]
if X_test is not None:
X_test = X_test.iloc[:, idxs_selected]
if X_test is not None:
# print(X_train.describe())
# print(X_test.describe())
return X_train, y_train, X_test
return X_train, y_train
print(f'Val loss: {val_loss/len(val_loader):.4f}',
f'Val acc:{val_total_acc/len(labels):.4f}')
return val_loss
print('Stage1: load data')
# 读取训练集,测试集和验证集
train = np.load('../train/10type_sort_train_data_8192.npy')
val = np.load('../val/10type_sort_eval_data_8192.npy')
# 读取训练集和验证集的标签,测试集是没有标签的,需要你使用模型进行分类,并将结果进行提交
train_label = np.load('../train/10type_sort_train_label_8192.npy')
val_label = np.load('../val/10type_sort_eval_label_8192.npy')
print('Stage2: data over_sampling')
smote = SMOTE(random_state=42, n_jobs=-1)
x_train, y_train = smote.fit_resample(train, train_label)
train_sp = get_fft_and_scaler(x_train, start=6892, end=7192)
val_sp = get_fft_and_scaler(val, start=6892, end=7192)
# 将数据转换成pytorch的tensor
print('Stage3: transform numpy data to tensor')
batch_size = 128
train_tensor = torch.tensor(train_sp).float()
y_train_tensor = torch.tensor(y_train).long()
val_tensor = torch.tensor(val_sp).float()
y_val_tensor = torch.tensor(val_label).long()
# 使用Dataloader对数据进行封装
("num", num_pipeline, num_col),
("one", OneHotEncoder(), cat_col_one),
#("ord", OneHotEncoder(), cat_col_ord),
])
features = full_pipeline.fit_transform(x)
#Ok, so now we have a training set encoded...
#It is very imblanced.. so we need to correct this, else have issues finding a good classification
from imblearn.over_sampling import SMOTE
from imblearn.over_sampling import RandomOverSampler
from imblearn.under_sampling import RandomUnderSampler
rus = RandomUnderSampler(random_state=0)
sm = SMOTE(ratio='auto', kind='regular')
ros = RandomOverSampler(random_state=0)
#X_train, y_train = sm.fit_sample(features,y)
X_train, y_train = ros.fit_sample(features,y)
#X_train, y_train = rus.fit_sample(features,y)
#X_train = features
#y_train = y
#Finally, we have 'feat' and a target 'Y' we can begin modeling
#Prep test data
xt = test.drop(target, axis=1)
y_test = test[target].copy()
labels.values.ravel(),
train_size=train_size,
shuffle=True,
stratify=labels.values.ravel())
# ### Impute Data
if data_impute:
imp = IterativeImputer(max_iter=25, random_state=1337)
X_train = imp.fit_transform(X_train)
X_test = imp.transform(X_test)
# ### Augment Data
if smote_ratio > 0:
smote = SMOTE(sampling_strategy='all',
random_state=1337,
k_neighbors=5,
n_jobs=1)
X_train, y_train = smote.fit_resample(X_train, y_train)
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
# ## Define Model
knn = KNeighborsClassifier(
n_neighbors=5,
weights='uniform', # or distance
p=2,
n_jobs=8)
if __name__ == '__main__':
# Load data
df1 = pd.read_csv('experiment.csv')
df2 = pd.read_csv('literature.csv')
columns = ['M', 'M/CTA', 'PC/M', 'time', 'Mn', 'MWD', 'others']
experiment = df1[columns]
literature = df2[columns]
# Over sampling
dataset_oversampling = pd.DataFrame()
for name, group in literature.groupby('others'):
temp = pd.concat([experiment, group])
temp_dummies = pd.get_dummies(temp)
smo = SMOTE()
X_smo, y_smo = smo.fit_sample(temp_dummies, temp_dummies['others_A'])
X_smo = pd.DataFrame(X_smo, columns=temp_dummies.columns)
dataset_oversampling = pd.concat([dataset_oversampling, X_smo])
dataset_oversampling = dataset_oversampling.fillna(0)
dataset_oversampling = dataset_oversampling.drop_duplicates()
dataset_oversampling.to_csv('dataset oversampling.csv',
index=False,
sep=',')
# Read the modified dataset
y = dataset_oversampling['Mn']
X = dataset_oversampling.drop(['Mn', 'MWD'], axis=1)
# Split the data into training and test sets
# Glucose: Generate a log feature and keep the original feature
dataset['ln_glucose_lvl'] = np.log(dataset['avg_glucose_level'])
smokes_df = pd.get_dummies(dataset['smoking_status'], 'smoke', drop_first=True)
dataset = dataset.drop('smoking_status', axis=1)
dataset = pd.concat([dataset, smokes_df], axis=1)
print(dataset.head())
k, seed = 1, 42
X = dataset.drop('stroke', axis=1)
y = dataset.stroke
# increases variety of training samples
sm = SMOTE(sampling_strategy='auto', k_neighbors=k, random_state=seed)
X_res, y_res = sm.fit_resample(X, y)
# Splits data into train, validation, and test
X_train, X_valid, y_train, y_valid = train_test_split(X_res, y_res, test_size=0.2)
X_train, X_test, y_train, y_test = train_test_split(X_train, y_train, test_size=0.25)
searched_params = {'subsample': 0.8, 'min_child_weight': 1, 'max_depth': 7, 'learning_rate': 0.25,
'grow_policy': 'lossguide', 'gamma': 1.5, 'colsample_bytree': 0.6}
xgbc = XGBClassifier(**searched_params, use_label_encoder=False)
X_train = np.concatenate([X_train, X_valid], axis=0)
y_train = np.concatenate([y_train, y_valid], axis=0)
xgbc.fit(X_train, y_train)
# final_preds = xgbc.predict(X_test)
def run(inputFile, n_trees, m_nodes, random_s, epochs, folds, kneighbors,
cores):
"""
Random Forest 主程序
参数
----
inputFile: 训练集文件路径
n_tress: 树大小
m_nodes: 最大节点数
random_s: 随机种子
epochs: 每个fold的训练次数
folds: k-fold折数
kneighbors: k邻近点数
cores: CPU核心数
"""
# The number of cores per socket in the machine
NUM_PARALLEL_EXEC_UNITS = cores
try:
# 导入训练数据
df = pd.read_csv(inputFile, encoding='utf8')
except (OSError) as e:
print("\n\t", e)
print(
"\nPlease make sure you input correct filename of training dataset!"
)
sys.exit(1)
# 分类矩阵为第一列数据(分类值减去1)
n_target = df.iloc[:, 0].values - 1
# 特征矩阵为去第一列之后数据
n_features = df.iloc[:, 1:].values
# 读取特征名称
features = df.columns[1:]
print("\nDataset shape: ", df.shape, " Number of features: ",
features.size)
# 不同 Class 样本数统计 (根据第1列)
np_target_y = np.asarray(np.unique(n_target, return_counts=True))
df_y = pd.DataFrame(np_target_y.T, columns=['Class', 'Sum'])
print("\nNumber of samples:\n{0}".format(df_y))
# 总样本数及分类数
num_samples = n_target.size
n_class = df_y['Class'].size
# Apply SMOTE 生成 fake data
sm = SMOTE(k_neighbors=kneighbors)
x_resampled, y_resampled = sm.fit_sample(n_features, n_target)
# after over sampleing 读取分类信息并返回数量
np_resampled_y = np.asarray(np.unique(y_resampled, return_counts=True))
df_resampled_y = pd.DataFrame(np_resampled_y.T, columns=['Class', 'Sum'])
print("\nNumber of samples after over sampleing:\n{0}\n".format(
df_resampled_y))
# 模型参数
num_steps = epochs # Total steps to train
num_features = features.size
num_trees = n_trees
max_nodes = m_nodes
# 设定 K-fold 分割器
rs = KFold(n_splits=folds, shuffle=True, random_state=random_s)
# Input and Target data
X = tf.placeholder(tf.float32, shape=[None, num_features])
# For random forest, labels must be integers (the class id)
Y = tf.placeholder(tf.int32, shape=[None])
# Random Forest Parameters
hparams = tensor_forest.ForestHParams(num_classes=n_class,
num_features=num_features,
num_trees=num_trees,
max_nodes=max_nodes).fill()
# Build the Random Forest
forest_graph = tensor_forest.RandomForestGraphs(hparams)
# print(forest_graph.params.__dict__)
# Get training graph and loss
train_op = forest_graph.training_graph(X, Y)
loss_op = forest_graph.training_loss(X, Y)
# Measure the accuracy
infer_op, _, _ = forest_graph.inference_graph(X)
pred_val = tf.argmax(infer_op, 1)
correct_prediction = tf.equal(pred_val, tf.cast(Y, tf.int64))
accuracy_op = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# Initialize the variables (i.e. assign their default value)
# init = tf.global_variables_initializer()
# Initialize the variables (i.e. assign their default value) and forest resources
init_vars = tf.group(
tf.global_variables_initializer(),
resources.initialize_resources(resources.shared_resources()))
# 生成 k-fold 索引
resampled_index_set = rs.split(y_resampled)
# 初始化折数
k_fold_step = 1
# 暂存每次选中的测试集和对应预测结果
test_cache = pred_cache = np.array([], dtype=np.int)
# Change the parallelism threads and OpenMP* make use of all the cores available in the machine.
# https://software.intel.com/en-us/articles/tips-to-improve-performance-for-popular-deep-learning-frameworks-on-multi-core-cpus
config = tf.ConfigProto(
intra_op_parallelism_threads=NUM_PARALLEL_EXEC_UNITS,
inter_op_parallelism_threads=2,
allow_soft_placement=True,
device_count={'CPU': NUM_PARALLEL_EXEC_UNITS})
os.environ["OMP_NUM_THREADS"] = "NUM_PARALLEL_EXEC_UNITS"
os.environ["KMP_BLOCKTIME"] = "30"
os.environ["KMP_SETTINGS"] = "1"
os.environ["KMP_AFFINITY"] = "granularity=fine,verbose,compact,1,0"
# 迭代训练 k-fold 交叉验证
for train_index, test_index in resampled_index_set:
# Start TensorFlow session
# sess = tf.train.MonitoredSession()
sess = tf.Session(config=config)
# Set random seed
tf.set_random_seed(random_s)
# Run the initializer
sess.run(init_vars)
print("\nFold:", k_fold_step)
# Training
for i in range(1, num_steps + 1):
# Prepare Data
batch_x = x_resampled[train_index] # 特征数据用于训练
batch_y = y_resampled[train_index] # 标记结果用于验证
batch_size = train_index.shape[0]
_, l = sess.run([train_op, loss_op],
feed_dict={
X: batch_x,
Y: batch_y
})
# 输出训练结果
if i % DISPLAY_STEP == 0 or i == 1:
acc = sess.run(accuracy_op, feed_dict={X: batch_x, Y: batch_y})
print("\nTraining Epoch:", '%06d' % i, "Train Accuracy:",
"{:.6f}".format(acc), "Train Loss:", "{:.6f}".format(l),
"Train Size:", batch_size)
# 输入测试数据
# 验证测试集 (通过 index 去除 fake data)
real_test_index = test_index[test_index < num_samples]
batch_test_x = x_resampled[real_test_index]
batch_test_y = y_resampled[real_test_index]
batch_test_size = len(real_test_index)
# 代入TensorFlow计算图验证测试集
accTest, costTest, predVal = sess.run([accuracy_op, loss_op, pred_val],
feed_dict={
X: batch_test_x,
Y: batch_test_y
})
# print("\n", predVal)
print("\nFold:", k_fold_step, "Test Accuracy:",
"{:.6f}".format(accTest), "Test Loss:",
"{:.6f}".format(costTest), "Test Size:", batch_test_size)
# 暂存每次选中的测试集和预测结果
test_cache = np.concatenate((test_cache, batch_test_y))
pred_cache = np.concatenate((pred_cache, predVal))
print(
"\n========================================================================="
)
# 每个fold训练结束后次数 +1
k_fold_step += 1
# 模型评估结果输出
from .utils import model_evaluation
model_evaluation(n_class, test_cache, pred_cache)
'alcohol': 'ALCEVER',
'marijuana': 'MJEVER',
'opioid': 'OPEVER',
'nicotine': 'NICEVR'
}
for i in ['alcohol', 'marijuana', 'opioid', 'nicotine']:
filename = '/Users/andyliu/Documents/LR_' + i + '.csv'
df = pd.read_csv(filename, header=0)
df = df.dropna()
X = df.loc[:, df.columns != dic[i]]
y = df.loc[:, df.columns == dic[i]].astype(np.int16)
os = SMOTE(random_state=0)
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.3,
random_state=0)
columns = X_train.columns
os_data_X, os_data_y = os.fit_sample(X_train, y_train.values.ravel())
os_data_X = pd.DataFrame(data=os_data_X, columns=columns)
os_data_y = pd.DataFrame(data=os_data_y, columns=[dic[i]])
logreg = LogisticRegression(solver='lbfgs')
rfe = RFE(logreg, 15)
rfe = rfe.fit(os_data_X, os_data_y.values.ravel())
print(i + " results:")
rfe_approved = (rfe.support_).tolist()
rfe_indices = []
def overSamplingSMOTE(dataset):
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=27)
X_train, y_train = sm.fit_sample(dataset[:,:-1], dataset[:,-1])
print(y_train)
from bayes_opt import BayesianOptimization
# 读取数据
data = pd.read_csv('../data/批发和零售业_处理.csv' , engine = "python")
data2 = pd.read_csv('../data/批发和零售业_处理.csv',engine = "python")
# print("data:\n")
# print(data)
data2.drop(data2.columns[[0]],axis = 1,inplace = True)
# print("data2删除第一列:\n")
# print(data2)
data2.drop(columns = ['FLAG'],axis = 1,inplace = True)
# print("data2删除FLAG:\n")
# print(data2)
# print("data:\n")
# print(data)
#定义SMOTE模型,random_state相当于随机数种子的作用
smo = SMOTE(sampling_strategy={1:45,0:789 },random_state=42)
# ros = RandomOverSampler(random_state=30,sampling_strategy=0.02)
X_smo,y_smo = smo.fit_resample(data2.iloc[:,:],data['FLAG'])
#标准化数据
sc=StandardScaler()
sc.fit(X_smo)#计算样本的均值和标准差
X_smo=pd.DataFrame(sc.transform(X_smo))
#拆分专家样本集
data_tr, data_te, label_tr, label_te = train_test_split(X_smo,y_smo,test_size=0.3)
print(label_tr.groupby(label_tr).count())
# 拆分专家样本集
# data_tr, data_te, label_tr, label_te = train_test_split(X_resampled, y_resampled,random_state=10,test_size=0.3)
# print("data_tr:\n")
# print(data_tr)
from sklearn.preprocessing import LabelEncoder lenc = LabelEncoder() lenc.fit(df_train['Departure']) df_train['Departure'] = lenc.transform(df_train['Departure']) df_train['Arrival'] = lenc.transform(df_train['Arrival']) from sklearn.preprocessing import OneHotEncoder enc = OneHotEncoder(categorical_features = [0,3,8,9]) df_train= enc.fit_transform(df_train).toarray() from sklearn.preprocessing import MinMaxScaler sc = MinMaxScaler() df_train = sc.fit_transform(df_train) from sklearn.model_selection import train_test_split X_train , X_test , y_train , y_test = train_test_split(df_train , y_train , test_size = 0.25) y_train = np.ravel(y_train) from imblearn.over_sampling import SMOTE ovs = SMOTE() X_train_res , y_train_res = ovs.fit_sample(X_train , y_train) from sklearn.naive_bayes import MultinomialNB clf = MultinomialNB() clf.fit(X_train_res , y_train_res) y_pred = clf.predict(X_test) from sklearn.metrics import f1_score x = f1_score(y_test , y_pred , average='micro')
dgs1 = GridSearchCV(estimator=model1,
param_grid=dectree_param_grid,
scoring='accuracy',
cv=5,
verbose=1,
n_jobs=-1)
dgs1.fit(X_train, y_train)
cv1 = dgs1.predict(x_val)
print('dec tree acc =: ', accuracy_score(y_val, cv1))
print('dec tree f1 score= ', f1_score(y_val, cv1, average='micro'))
print('parameters=', dgs1.best_params_)
joblib.dump(dgs1, "1-dectree1.joblib")
acc.append(accuracy_score(y_val, cv1))
f1.append(f1_score(y_val, cv1, average='micro'))
model2 = imbpipe([('sampling', SMOTE()), ('scl', StandardScaler()),
('clf', DecisionTreeClassifier(random_state=101))])
dgs2 = GridSearchCV(estimator=model2,
param_grid=dectree_param_grid,
scoring='accuracy',
cv=5,
verbose=1,
n_jobs=-1)
dgs2.fit(X_train, y_train)
cv2 = dgs2.predict(x_val)
print('dec tree SMOTE acc =: ', accuracy_score(y_val, cv2))
print('dec tree f1 score= ', f1_score(y_val, cv2, average='micro'))
print('parameters=', dgs2.best_params_)
joblib.dump(dgs2, "2-dectree2.joblib")
np.load(os.path.join(read_data.tweet_representation_path, 'tweet_array_2017.npy'))
X_train_valid = np.load(
os.path.join(read_data.tweet_representation_path,
'train_valid_cross_validation_data.npy'))
y_train_valid = np.load(
os.path.join(read_data.tweet_representation_path,
'train_valid_cross_validation_label.npy'))
X_test = np.load(
os.path.join(read_data.tweet_representation_path,
'test_data_for_model_compare.npy'))
y_test = np.load(
os.path.join(read_data.tweet_representation_path,
'test_label_for_model_compare.npy'))
#
# Use SMOTE to do the oversampling
smt = SMOTE(random_state=random_seed, k_neighbors=1)
oversampled_train_validate_data, oversampled_train_validate_y = smt.fit_sample(
X_train_valid, y_train_valid)
print('====================================================')
print('The distribution of the train_valid_data is: ')
print(Counter(y_train_valid))
print('The distribution of the oversampled data is: ')
print(Counter(oversampled_train_validate_y))
print('====================================================')
# Build the Classifiers
ffnn_model = get_ffnn_model()
ffnn_model.summary()
# The KerasClassifier Wrapper helps us GridSearch the hyperparameters of our neural net
ffnn_model_wrapper = KerasClassifier(build_fn=get_ffnn_model,
verbose=0,
# data['n_follower'] = np.log1p(data['n_follower'])
data['n_fan'] = np.log1p(data['n_fan'])
data['n_weibo'] = np.log1p(data['n_weibo'])
data['gap_avg'] = np.log1p(data['gap_avg'])
data['gap_min'] = np.log1p(data['gap_min'])
data['gap_max'] = np.log1p(data['gap_max'])
data['n_gap_less_mean'] = np.log1p(data['n_gap_less_mean'])
data['n_gap_less_60'] = np.log1p(data['n_gap_less_60'])
data = shuffle(data)
y = data['class'].ravel()
x = data.drop(['class'], axis=1)
# balance positive and negative data
print('Before balancing: 0:1 = %s:%s' % (np.sum(y == 0), np.sum(y == 1)))
sm = SMOTE(kind='regular')
x_res, y_res = sm.fit_sample(x, y)
print('After balancing: 0:1 = %s:%s' %
(np.sum(y_res == 0), np.sum(y_res == 1)))
# split train and test data
x_train, x_test, y_train, y_test = train_test_split(x_res,
y_res,
test_size=0.2)
n_train = x_train.shape[0]
n_test = x_test.shape[0]
# some variables
SEED = 0 # for reproducibility
NFOLDS = 5 # set folds for out-of-fold prediction
def split_resample(X, y, normalize=True, resample=True, verbose=True):
'''
Creates train-test-splits, normalizes continuous features, resamples training
data using SMOTE.
Input:
- X: pandas df or numpy array (features)
- y: pandas series or numpy array (label)
- normalize: boolean indicating whether to normalize continuous features
- resample: boolean indicating whether to resample minority classes in training data
Returns:
- X_train, X_test, y_train, y_test
- Also writes X_train, X_test, y_train, y_test to local CSV files
'''
start_time = time.time()
# Split into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.2,
random_state=0)
# Normalize continuous features based on training set
if normalize:
scaler = StandardScaler()
X_train[CONTINUOUS_COLUMNS] = scaler.fit_transform(
X_train[CONTINUOUS_COLUMNS])
X_test[CONTINUOUS_COLUMNS] = scaler.transform(
X_test[CONTINUOUS_COLUMNS])
# Resample training data to balance classes
if resample:
# Under-sample majority classes
under_sampling_dict = {
'Closed with explanation':
int(get_count(y_train, 'Closed with explanation') * FRAC_MAJORITY),
'Closed':
get_count(y_train, 'Closed'),
'Untimely response':
get_count(y_train, 'Untimely response'),
'Closed with non-monetary relief':
get_count(y_train, 'Closed with non-monetary relief'),
'Closed with monetary relief':
get_count(y_train, 'Closed with monetary relief'),
}
rus = RandomUnderSampler(sampling_strategy=under_sampling_dict,
random_state=0)
X_train, y_train = rus.fit_resample(X_train, y_train)
# Over-sample minority classes
sm = SMOTE(random_state=0)
X_train, y_train = sm.fit_sample(X_train, y_train)
# Write to csv
X_train.to_csv(os.path.join(PROCESSED_DATA_DIR, 'X_train.csv'))
y_train.to_csv(os.path.join(PROCESSED_DATA_DIR, 'y_train.csv'))
X_test.to_csv(os.path.join(PROCESSED_DATA_DIR, 'X_test.csv'))
y_test.to_csv(os.path.join(PROCESSED_DATA_DIR, 'y_test.csv'))
time_elapsed = time.time() - start_time
if verbose:
print('Distribution of training labels: \n{}'.format(
y_train.value_counts() / y_train.shape[0]))
print('\n')
print('Training samples:', X_train.shape[0])
print('Testing samples:', X_test.shape[0])
print('Time elapsed:', time_elapsed)
return X_train, X_test, y_train, y_test
# <h2 id="t10" style="margin-bottom: 18px">Over-sampling: SMOTE</h2> # # SMOTE (Synthetic Minority Oversampling TEchnique) consists of synthesizing elements for the minority class, based on those that already exist. It works randomly picingk a point from the minority class and computing the k-nearest neighbors for this point. The synthetic points are added between the chosen point and its neighbors. # 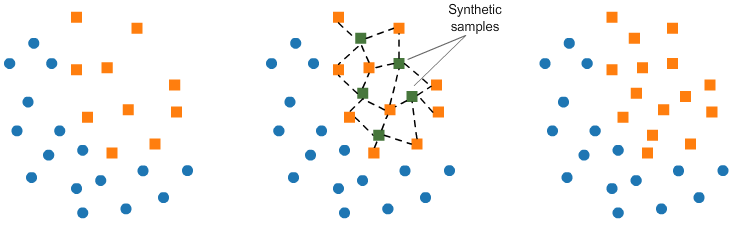 # We'll use <code>ratio='minority'</code> to resample the minority class. # In[ ]: from imblearn.over_sampling import SMOTE smote = SMOTE(ratio='minority') X_sm, y_sm = smote.fit_sample(X, y) plot_2d_space(X_sm, y_sm, 'SMOTE over-sampling') # <h2 id="t11" style="margin-bottom: 18px">Over-sampling followed by under-sampling</h2> # # Now, we will do a combination of over-sampling and under-sampling, using the SMOTE and Tomek links techniques: # In[ ]: from imblearn.combine import SMOTETomek smt = SMOTETomek(ratio='auto')
#!pip install imblearn #encode categorical variable #from sklearn.preprocessing import LabelEncoder #encoder = LabelEncoder() #x_train.record = encoder.fit_transform(x_train.record) #x_test.record = encoder.transform(x_test.record) #the encoded feature x_train.record #There is still an imbalance in the class distribution. For this, we use SMOTE only on the training data to handle this. import imblearn from imblearn.over_sampling import SMOTE smote = SMOTE(random_state=1) x_train_balanced, y_balanced = smote.fit_sample(x_train, y_train) y_train.value_counts() #min max scaler from sklearn.preprocessing import MinMaxScaler scaler = MinMaxScaler() normalised_train_df = scaler.fit_transform(x_train_balanced.drop(columns=['record'])) normalised_train_df = pd.DataFrame(normalised_train_df, columns=x_train_balanced.drop(columns=['record']).columns) normalised_train_df['record'] = x_train_balanced['record'] x_test = x_test.reset_index(drop=True) normalised_test_df = scaler.transform(x_test.drop(columns=['record'])) normalised_test_df = pd.DataFrame(normalised_test_df, columns=x_test.drop(columns=['record']).columns)
def prepare_model_data(df, y_col, drop_cols = [],
test_size = .3, scale_x = True, scale_y = False,
oversample = True):
"""
Purpose: This function prepares the data to be modeled by conducting a train/test split and standaridizng the data (subtracting mean dividing by standard deviation) if specified
Input:
* df = data frame with predictor variables and variable of interest
* y_col = column name for variable of interest
* drop_cols = variables that do not need to be included in the predictor variables (not including the variable of interest)
* test_size = the proportion of the data frame to isolate for testing
- default = 30%
* scale_x = whether to scale the X variables with a StandardScaler
* scale_y = whether to scale the y variable with a StandardScaler
* oversample = whether to oversample the training set
Output:
* X_train: data frame with predictor variables to train model on
* X_test: data frame with predictor variables to test model on
* y_train: data frame with actual values of variable of interest for train set
* y_test: data frame with actual values of variable of interest for test set
* scalers: dictionary with fit scalers (empty if not applicable)
"""
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from imblearn.over_sampling import SMOTE
import matplotlib.pyplot as plt
drop_cols.append(y_col)
X = df.drop(drop_cols, axis='columns')
y = df[[y_col]]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = test_size)
column_names = X_train.columns
X_train, X_test, y_train, y_test = X_train.values, X_test.values, \
y_train.values.ravel(), y_test.values.ravel()
def plot_y(y):
index = [0, 1]
values = [sum(y == 0), sum(y == 1)]
plt.bar(index, values)
plt.ylabel('Count')
plt.title(f'Distribution of {y_col}');
if oversample:
n = sum(y_train == 1)
y_before = y_train.copy()
print(f'Before oversampling, the minor class of the traing set has {n} samples.')
oversample = SMOTE()
X_train, y_train = oversample.fit_resample(X_train, y_train)
n = sum(y_train == 1)
print(f'After oversampling, the minor class of the traing set has {n} samples.')
plt.figure(figsize=(8,4))
plt.subplot(1,2,1)
plot_y(y_before)
plt.subplot(1,2,2)
plot_y(y_train)
plt.tight_layout()
scalers = {}
if scale_x:
scaler_x = StandardScaler().fit(X_train)
X_train = X_train.copy()
X_train = scaler_x.transform(X_train)
X_test = X_test.copy()
X_test= scaler_x.transform(X_test)
scalers['X'] = scaler_x
if scale_y:
scaler_y = StandardScaler().fit(y_train.reshape(-1,1))
y_train = y_train.copy()
y_train = scaler_y.transform(y_train.reshape(-1,1)).ravel()
y_test = y_test.copy()
y_test = scaler_y.transform(y_test.reshape(-1,1)).ravel()
scalers['y'] = scaler_y
return X_train, X_test, y_train, y_test, scalers, column_names
from sklearn.ensemble import RandomForestClassifier
from imblearn.over_sampling import SMOTE, RandomOverSampler
from prepare import readbunchobj
import pandas as pd
from sklearn.metrics import fbeta_score, make_scorer
from sklearn import metrics
ftwo_scorer = make_scorer(fbeta_score, beta=2)
X = pd.read_excel('X.xlsx')
y = pd.read_excel('y.xlsx')
# X_smote, y_smote = SMOTE().fit_resample(X, y)
X_r, y_r = RandomOverSampler().fit_sample(X, y.values.ravel())
X_s, y_s = SMOTE().fit_sample(X, y.values.ravel())
# # 调参
# print('Start adjusting parameters')
# param_test1 = {'n_estimators': range(10, 200, 10)}
#
# param_test2 = {'max_depth': range(3, 30, 5),
# 'min_samples_split': range(50, 201, 20)}
#
# param_test3 = {'min_samples_split': range(10, 81, 10),
# 'min_samples_leaf': range(10, 60, 10)}
#
# param_test4 = {'max_features': range(3, 83, 5)}
#
#
# gsearch1 = GridSearchCV(estimator=
model_evaluation(y_test, pred)
# SMOTE을 이용해서 Oversampling을 진행해보자!
# 기존의 X_train, y_train, X_test, y_test의 형태 확인
print("Number transactions X_train dataset: ", X_train.shape)
print("Number transactions y_train dataset: ", y_train.shape)
print("Number transactions X_test dataset: ", X_test.shape)
print("Number transactions y_test dataset: ", y_test.shape)
from imblearn.over_sampling import SMOTE
print("Before OverSampling, counts of label '1': {}".format(sum(y_train == 1))) # y_train 중 레이블 값이 1인 데이터의 개수
print("Before OverSampling, counts of label '0': {} \n".format(sum(y_train == 0))) # y_train 중 레이블 값이 0 인 데이터의 개수
sm = SMOTE(random_state = 42, ratio = 0.3) # SMOTE 알고리즘, 비율 증가
X_train_res, y_train_res = sm.fit_sample(X_train, y_train.ravel()) # Over Sampling 진행
print("After OverSampling, counts of label '1': {}".format(sum(y_train_res==1)))
print("After OverSampling, counts of label '0': {}".format(sum(y_train_res==0)))
print("Before OverSampling, the shape of X_train: {}".format(X_train.shape)) # SMOTE 적용 이전 데이터 형태
print("Before OverSampling, the shape of y_train: {}".format(y_train.shape)) # SMOTE 적용 이전 데이터 형태
print('After OverSampling, the shape of X_train: {}'.format(X_train_res.shape)) # SMOTE 적용 결과 확인
print('After OverSampling, the shape of y_train: {}'.format(y_train_res.shape)) # # SMOTE 적용 결과 확인
lgb_dtrain2 = lgb.Dataset(data = pd.DataFrame(X_train_res), label = pd.DataFrame(y_train_res)) # 학습 데이터를 LightGBM 모델에 맞게 변환
lgb_param2 = {'max_depth': 10, # 트리 깊이
'learning_rate': 0.01, # Step Size
