def get_binary_Tomek_Links_cleaned_data(id_df, X_df, y_df):
tLinks = TomekLinks()
a = y_df.iloc[:, 0]
tLinks.fit_sample(X_df, y_df.iloc[:, 0])
sample_indices = tLinks.sample_indices_
id_df_cleaned = id_df.iloc[sample_indices]
X_df_cleaned = X_df.iloc[sample_indices]
y_df_cleaned = y_df.iloc[sample_indices]
return id_df_cleaned, X_df_cleaned, y_df_cleaned
def getData(splitData=True, useImbalancer=False, useStratify=False):
global standard_scaler
data = pd.read_csv(filepath_or_buffer="DataSource/binary.csv")
X = data.values[:, 1:-1]
rank_dummy = pd.get_dummies(data['rank'], drop_first=True).to_numpy()
X = np.concatenate((X, rank_dummy), axis=1)
y = data.values[:, 0].reshape(-1, 1)
if useStratify:
stratify = y
else:
stratify = None
if splitData:
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.2,
random_state=101,
shuffle=True,
stratify=stratify)
else:
X_train = X
y_train = y
if useImbalancer and splitData:
tl = TomekLinks(sampling_strategy='majority')
X_train, y_train = tl.fit_sample(X=X_train, y=y_train)
# print("After 1st pass: "******"After 2nd pass: "******"After 3rd pass: "******"After 4th pass: "******"After 5th pass: "******"After 6th pass: "******"y_train\n", np.asarray((unique, counts)).T)
if splitData:
unique, counts = np.unique(y_test, return_counts=True)
# print("y_test\n", np.asarray((unique, counts)).T)
if splitData:
return X_train, X_test, y_train.ravel(), y_test.ravel()
else:
return X_train, y_train.ravel()
def undersample(X, y, bal_strategy): print 'Shape of X: ', X.shape print 'Shape of y_Train: ', y.shape if(bal_strategy == "RANDOM" or bal_strategy == "ALL"): # apply random under-sampling rus = RandomUnderSampler() X_sampled, y_sampled = rus.fit_sample(X, y) print 'Shape of X_sampled: ', X_sampled.shape print 'Shape of y_sampled: ', y_sampled.shape elif(bal_strategy == "TOMEK" or bal_strategy == "ALL"): # Apply Tomek Links cleaning tl = TomekLinks() X_sampled, y_sampled = tl.fit_sample(X, y) print 'Shape of X_sampled: ', X_sampled.shape print 'Shape of y_sampled: ', y_sampled.shape elif(bal_strategy == 'NONE'): X_sampled = X y_sampled = y print 'Shape of X_sampled: ', X_sampled.shape print 'Shape of y_sampled: ', y_sampled.shape else: print 'bal_stragegy not in ALL, RANDOM, TOMEK, NONE' sys.exit(1) return (X_sampled, y_sampled)
def undersample_tomek_link(X,
y,
label='Tomek links under-sampling',
plot=False):
tl = TomekLinks(return_indices=True, ratio='all')
X_tl, y_tl, id_tl = tl.fit_sample(X, y)
X_tl = pd.DataFrame(X_tl, columns=X.columns)
y_tl = pd.Series(y_tl, name=y.name)
if plot == True:
#print('Removed indexes:', id_tl)
# plotting using pca
pca = PCA(n_components=2)
X_pca = pd.DataFrame(pca.fit_transform(X_tl))
colors = ['#1F77B4', '#FF7F0E']
markers = ['o', 's']
for l, c, m in zip(np.unique(y_tl), colors, markers):
plt.scatter(
X_pca.loc[y_tl == l, 0], # pc 1
X_pca.loc[y_tl == l, 1], # pc 2
c=c,
label=l,
marker=m)
plt.title(label)
plt.legend(loc='upper right')
plt.show()
return X_tl, y_tl, tl, id_tl
def test_tl_fit_sample():
"""Test the fit sample routine"""
# Resample the data
tl = TomekLinks(random_state=RND_SEED)
X_resampled, y_resampled = tl.fit_sample(X, Y)
X_gt = np.array([[0.31230513, 0.1216318],
[0.68481731, 0.51935141],
[1.34192108, -0.13367336],
[0.62366841, -0.21312976],
[1.61091956, -0.40283504],
[-0.37162401, -2.19400981],
[0.74680821, 1.63827342],
[0.2184254, 0.24299982],
[0.61472253, -0.82309052],
[0.19893132, -0.47761769],
[0.97407872, 0.44454207],
[1.40301027, -0.83648734],
[-1.20515198, -1.02689695],
[-0.23374509, 0.18370049],
[-0.32635887, -0.29299653],
[-0.00288378, 0.84259929],
[1.79580611, -0.02219234]])
y_gt = np.array([1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0])
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
def test_tl_fit_sample():
"""Test the fit sample routine"""
# Resample the data
tl = TomekLinks(random_state=RND_SEED)
X_resampled, y_resampled = tl.fit_sample(X, Y)
X_gt = np.array([[0.31230513, 0.1216318],
[0.68481731, 0.51935141],
[1.34192108, -0.13367336],
[0.62366841, -0.21312976],
[1.61091956, -0.40283504],
[-0.37162401, -2.19400981],
[0.74680821, 1.63827342],
[0.2184254, 0.24299982],
[0.61472253, -0.82309052],
[0.19893132, -0.47761769],
[0.97407872, 0.44454207],
[1.40301027, -0.83648734],
[-1.20515198, -1.02689695],
[-0.23374509, 0.18370049],
[-0.32635887, -0.29299653],
[-0.00288378, 0.84259929],
[1.79580611, -0.02219234]])
y_gt = np.array([1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0])
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
def _tomek_data(self):
"""Performs tomek links. Can not handle nominal values."""
if self.cols_nominal.size > 0:
print("Skipping Tomek Links. Cannot perform with raw categorical data. Create dummies to use.")
return
tl = TomekLinks()
self.X_train, self.y_train = tl.fit_sample(self.X_train, self.y_train)
def get_tomeklinks_under_sampled_dataset():
tl = TomekLinks(return_indices=True, ratio='majority')
X_tl, y_tl, id_tl = tl.fit_sample(X_train, y_train)
print('Removed indexes:', id_tl)
shuffle(X_tl)
y_tl = X_tl[target]
return X_tl, y_tl
def test_tl_fit_sample():
"""Test the fit sample routine"""
# Resample the data
tl = TomekLinks(random_state=RND_SEED)
X_resampled, y_resampled = tl.fit_sample(X, Y)
currdir = os.path.dirname(os.path.abspath(__file__))
X_gt = np.load(os.path.join(currdir, 'data', 'tl_x.npy'))
y_gt = np.load(os.path.join(currdir, 'data', 'tl_y.npy'))
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
def imbalanced_resampling(method, x, y):
if method == "under":
sampling = TomekLinks(sampling_strategy="auto")
elif method == "over":
sampling = SMOTE(ratio='auto')
elif method == "combined":
sampling = SMOTETomek()
else:
return x, y
X, Y = sampling.fit_sample(x, y)
return X, Y
def test_tl_fit_sample():
"""Test the fit sample routine"""
# Resample the data
tl = TomekLinks(random_state=RND_SEED)
X_resampled, y_resampled = tl.fit_sample(X, Y)
currdir = os.path.dirname(os.path.abspath(__file__))
X_gt = np.load(os.path.join(currdir, 'data', 'tl_x.npy'))
y_gt = np.load(os.path.join(currdir, 'data', 'tl_y.npy'))
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
def oversample(self, train, labels):
"""
Over samples data according to SMOTE algorithm
"""
#Oversample
sm = SMOTE(random_state=2)
train_res, labels_res = sm.fit_sample(train, labels)
#clear noise points that emerged from oversampling
tl = TomekLinks(random_state=42)
train_res, labels_res = tl.fit_sample(train_res, labels_res)
return train_res, labels_res
def trainModelWithResults(model, X, y,rd_state=None,autoscale=1,usetomeklinks=1):
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size=0.25, stratify=y, random_state=rd_state) # stratify the split because we have unbalanced target
if autoscale==1:
scaler = StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
if usetomeklinks==1:
tl = TomekLinks(return_indices=False)
X_train, y_train = tl.fit_sample(X_train, y_train)
mfitted = model.fit(X_train,y_train)
predictions = mfitted.predict(X_test)
print(confusion_matrix(y_test, predictions))
print(classification_report(y_test, predictions))
def test_tl_fit_sample_with_indices():
"""Test the fit sample routine with indices support"""
# Resample the data
tl = TomekLinks(return_indices=True, random_state=RND_SEED)
X_resampled, y_resampled, idx_under = tl.fit_sample(X, Y)
currdir = os.path.dirname(os.path.abspath(__file__))
X_gt = np.load(os.path.join(currdir, 'data', 'tl_x.npy'))
y_gt = np.load(os.path.join(currdir, 'data', 'tl_y.npy'))
idx_gt = np.load(os.path.join(currdir, 'data', 'tl_idx.npy'))
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
assert_array_equal(idx_under, idx_gt)
def test_tl_fit_sample_with_indices():
"""Test the fit sample routine with indices support"""
# Resample the data
tl = TomekLinks(return_indices=True, random_state=RND_SEED)
X_resampled, y_resampled, idx_under = tl.fit_sample(X, Y)
currdir = os.path.dirname(os.path.abspath(__file__))
X_gt = np.load(os.path.join(currdir, 'data', 'tl_x.npy'))
y_gt = np.load(os.path.join(currdir, 'data', 'tl_y.npy'))
idx_gt = np.load(os.path.join(currdir, 'data', 'tl_idx.npy'))
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
assert_array_equal(idx_under, idx_gt)
def dataset_sampling(X,y):
sm = SMOTE(random_state=42,ratio='minority')
smt = SMOTETomek(ratio='auto')
ros = RandomOverSampler(random_state=0)
rus = RandomUnderSampler(random_state=0)
tl = TomekLinks(return_indices=True, ratio='majority')
cc = ClusterCentroids(ratio={0: 10})
#X_res, y_res = sm.fit_resample(X, y)
#X_res, y_res = ros.fit_resample(X, y)
#X_res, y_res = rus.fit_resample(X, y)
X_res, y_res, id_tl = tl.fit_sample(X, y)
#X_res, y_res = cc.fit_sample(X, y)
#X_res, y_res = smt.fit_sample(X, y)
return X_res,y_res
def undersampling(df):
tl = TomekLinks(ratio='all', n_jobs=16, return_indices=True)
X = []
y = []
add2list(df, X, y)
X, y, idx = tl.fit_sample(X, y)
criterion = [False] * len(df)
for i in idx:
criterion[i] = True
newdf = df[criterion].reset_index(drop=True)
return newdf
def resample(X, y, method):
if method == 'smote':
sm = SMOTE(random_state=2777, ratio=1.0)
X, y = sm.fit_sample(X, y)
elif method == 'tomek':
tomek = TomekLinks(random_state=2777, sampling_strategy='majority')
X, y = tomek.fit_sample(X, y)
elif method == 'smote-tomek':
smt = SMOTETomek(random_state=2777, ratio='auto')
X, y = smt.fit_sample(X, y)
elif method == 'none':
pass
else:
raise ValueError('Resampling method not recognized.')
return X, y
def test_tl_fit_sample_with_indices():
tl = TomekLinks(return_indices=True, random_state=RND_SEED)
X_resampled, y_resampled, idx_under = tl.fit_sample(X, Y)
X_gt = np.array([[0.31230513, 0.1216318], [0.68481731, 0.51935141],
[1.34192108, -0.13367336], [0.62366841, -0.21312976],
[1.61091956, -0.40283504], [-0.37162401, -2.19400981],
[0.74680821, 1.63827342], [0.2184254, 0.24299982],
[0.61472253, -0.82309052], [0.19893132, -0.47761769],
[0.97407872, 0.44454207], [1.40301027, -0.83648734],
[-1.20515198, -1.02689695], [-0.23374509, 0.18370049],
[-0.32635887, -0.29299653], [-0.00288378, 0.84259929],
[1.79580611, -0.02219234]])
y_gt = np.array([1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0])
idx_gt = np.array(
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 16, 17, 18, 19])
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
assert_array_equal(idx_under, idx_gt)
def test_tl_fit_sample_with_indices():
tl = TomekLinks(return_indices=True)
X_resampled, y_resampled, idx_under = tl.fit_sample(X, Y)
X_gt = np.array([[0.31230513, 0.1216318], [0.68481731, 0.51935141],
[1.34192108, -0.13367336], [0.62366841, -0.21312976],
[1.61091956, -0.40283504], [-0.37162401, -2.19400981],
[0.74680821, 1.63827342], [0.2184254, 0.24299982],
[0.61472253, -0.82309052], [0.19893132, -0.47761769],
[0.97407872, 0.44454207], [1.40301027, -0.83648734],
[-1.20515198, -1.02689695], [-0.23374509, 0.18370049],
[-0.32635887, -0.29299653], [-0.00288378, 0.84259929],
[1.79580611, -0.02219234]])
y_gt = np.array([1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0])
idx_gt = np.array(
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 16, 17, 18, 19])
assert_array_equal(X_resampled, X_gt)
assert_array_equal(y_resampled, y_gt)
assert_array_equal(idx_under, idx_gt)
def undersampling_pair(df1, df2):
tl = TomekLinks(ratio='auto', n_jobs=16, return_indices=True)
X = []
y = []
add2list(df1, X, y)
add2list(df2, X, y)
X, y, idx = tl.fit_sample(X, y)
l1 = len(df1)
l2 = len(df2)
criterion1 = [False] * l1
criterion2 = [False] * l2
for i in idx:
if i < l1:
criterion1[i] = True
else:
criterion2[i - l1] = True
newdf1 = df1[criterion1].reset_index(drop=True)
newdf2 = df2[criterion2].reset_index(drop=True)
return newdf1, newdf2
def under_sampling(self, x, y, method='random'):
# if the y data is dataframe, it must be flatted (num_sample, )
if isinstance(y, pd.DataFrame):
y_ = y.ravel()
else:
y_ = y
# use tomeklinks under-sample
if 'tomek' in str(method).lower():
tl = TomekLinks(return_indices=True, ratio='majority')
x_res, y_res, id_res = tl.fit_sample(x, y_)
# Need to be implemented
elif 'cluster' in str(method).lower():
pass
# the default option is use Random-Sample
else:
rus = RandomUnderSampler(return_indices=True)
x_res, y_res, id_res = rus.fit_sample(x, y_)
# return desired information
if self.return_indices:
return x_res, y_res, id_res
return x_res, y_res
def sampling(X_train, y_train):
ran_over = RandomOverSampler(random_state=42)
X_train_oversample,y_train_oversample = ran_over.fit_resample(X_train,y_train)
ran_under = RandomUnderSampler(random_state=42)
X_train_undersample, y_train_undersample = ran_under.fit_resample(X_train,y_train)
tl = TomekLinks(n_jobs=6)
X_train_tl, y_train_tl = tl.fit_sample(X_train, y_train)
sm = SMOTE(random_state=42, n_jobs=5)
X_train_sm, y_train_sm = sm.fit_sample(X_train, y_train)
enn = EditedNearestNeighbours()
X_train_enn, y_train_enn = enn.fit_resample(X_train, y_train)
print(np.unique(y_train, return_counts=True))
print("after sampling")
print("randomg over sampling")
print(np.unique(y_train_oversample, return_counts=True))
print("SMOTE sampling")
print(np.unique(y_train_sm, return_counts=True))
print("random under sampling")
print(np.unique(y_train_undersample, return_counts=True))
print("TomekLinks under sampling")
print(np.unique(y_train_tl, return_counts=True))
return (X_train_oversample, y_train_oversample, X_train_undersample, y_train_undersample,
X_train_tl, y_train_tl, X_train_sm, y_train_sm, X_train_enn, y_train_enn)
#
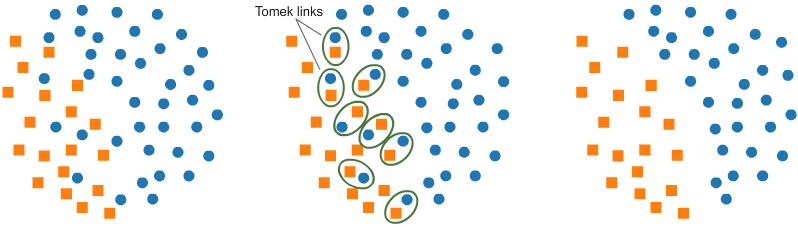
# - Elimination of closely intact data points from Majority Class increases the gap between the 2 classes which further eases the Classification Process.
#
#
# - Below Figure best explains the Tomek Link Technique.
# 
# In[23]:
#implementing tomek links
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(random_state=50, ratio='not minority')
x_tl_res, y_tl_res = tl.fit_sample(xtrain, ytrain)
print(ytrain.value_counts(), '\n')
np.bincount(y_tl_res)
# In[24]:
#applying logistic regression
est = LogisticRegression(solver='lbfgs')
est.fit(x_tl_res, y_tl_res)
pred = est.predict(xtest)
print('Prediction : ', pred, '\n')
print('Validation Score : ', est.score(xtest, ytest) * 100)
,predictions)
print(f'Recall Logistic Regression {recall: .2f}')
print(report)
print(balanced_accuracy_score(Y_test, predictions))
t1 = pl.time.time() - t0
print("Time taken: {:.0f} min {:.0f} secs".format(*divmod(t1, 60)))
print("best parameters",LR_model.best_params_)
plot_confusion_matrix(confusion_matrix(Y_test,predictions),['Dolphin','Non-Dolphin'])
# #forth test----------------------Logestic Regression--------------------------------with undersampling
from imblearn.under_sampling import TomekLinks
undersample = TomekLinks()
Xtrain_tomek, Ytrain_tomek = undersample.fit_sample(X_train, Y_train)
t0 = pl.time.time()
LR = LogisticRegression(max_iter=4000,
random_state=49,
n_jobs=1, class_weight='balanced') # for liblinear n_jobs is +1.
parameters = {"penalty": ['l1', 'l2'],'C': [0.001, 0.01, 0.1, 1, 10, 100, 1000], "solver":['liblinear','sag','saga']}
LR_model = GridSearchCV(LR, parameters, scoring="precision", cv=3)
# fit the classifier
LR_model.fit(Xtrain_tomek,Ytrain_tomek.values.ravel())
# get the prediction
predictions = LR_model.predict(X_test)
# samples. If ``ratio='auto'`` only the sample from the majority class will be
# removed. If ``ratio='all'`` both samples will be removed.
sampler = TomekLinks()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
ax_arr = (ax1, ax2)
title_arr = ('Removing only majority samples',
'Removing all samples')
for ax, title, sampler in zip(ax_arr,
title_arr,
[TomekLinks(ratio='auto', random_state=0),
TomekLinks(ratio='all', random_state=0)]):
X_res, y_res = sampler.fit_sample(np.vstack((X_minority, X_majority)),
np.array([0] * X_minority.shape[0] +
[1] * X_majority.shape[0]))
ax.scatter(X_res[y_res == 0][:, 0], X_res[y_res == 0][:, 1],
label='Minority class', s=200, marker='_')
ax.scatter(X_res[y_res == 1][:, 0], X_res[y_res == 1][:, 1],
label='Majority class', s=200, marker='+')
# highlight the samples of interest
ax.scatter([X_minority[-1, 0], X_majority[1, 0]],
[X_minority[-1, 1], X_majority[1, 1]],
label='Tomek link', s=200, alpha=0.3)
ax.set_title(title)
make_plot_despine(ax)
fig.tight_layout()
X_sampled2, y_sampled2 = smote.fit_sample(X, y)
# ROS와 SMOTE data unique성 비교
import pandas as pd
X_sampled1 = pd.DataFrame(X_sampled1)
len(X_sampled1.drop_duplicates()) # unique가 많지않다
X_sampled2 = pd.DataFrame(X_sampled2)
len(X_sampled2.drop_duplicates()) # new data가 있기 때문에 unique가 상대적으로 많긴 하다.
# Tomek Link
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(return_indices=True)
X_resampled, y_resampled, inds = tl.fit_sample(X, y)
# One-sided selection
# remove every data point, 근데 그 전에 k-nn을 적용해야한다.
from imblearn.under_sampling import OneSidedSelection
oss = OneSidedSelection(n_neighbors=1, n_seeds_S=1)
X_resampled, y_resampled = oss.fit_sample(X, y)
#Cost-sensitive Learning
svc = SVC(kernel='linear', class_weight={1: 10})
svc.fit(X, y)
y_pred = svc.predict(X)
from imblearn.under_sampling import TomekLinks
smote_nc = SMOTENC(categorical_features=[4, 5, 6, 7, 8, 9, 10, 11, 12],
random_state=0)
X_res, y_res = smote_nc.fit_resample(X_train, y_train)
geo_bool_mask = (X_res[:, 6] + X_res[:, 7] + X_res[:, 8]) == 1
X_res = X_res[geo_bool_mask]
y_res = y_res[geo_bool_mask]
gender_bool_mask = (X_res[:, 9] + X_res[:, 10]) == 1
X_resampled1 = X_res[gender_bool_mask]
y_resampled1 = y_res[gender_bool_mask]
print(sorted(Counter(y_resampled1).items()))
tl = TomekLinks()
X_resampled, y_resampled = tl.fit_sample(X_resampled1, y_resampled1)
svm_svc_poly_param_grid = {
'C': [0.01, 0.1, 1, 10, 100],
'gamma': [1e-6, 1e-5, 1e-4, 1e-3],
'kernel': ['poly'],
'degree': [2, 3, 4],
'class_weight': ['balanced', None],
'probability': [True, False],
'tol': [1e-5, 1e-4, 1e-3, 1e-2]
}
svm_svc_poly = SVC()
SVM_poly_grid_search = GridSearchCV(svm_svc_poly,
svm_svc_poly_param_grid,
cv=5,
refit=True,
plot_confusion_matrix(y_test, predict_mine) interact(update, var=FloatSlider(min=0.001, max=0.04, step=0.001)) # /Logistic Regression with balanced class weight to resolve the issue of imbalance data/ lr_b = LogisticRegression(max_iter=1000, class_weight='balanced') lr_b.fit(X_train, y_train) y_pred_b = lr_b.predict(X_test) plot_confusion_matrix(y_test, y_pred_b) # /Tomeklinks/ from imblearn.under_sampling import TomekLinks tl = TomekLinks(sampling_strategy='majority') X_train_tl, y_train_tl = tl.fit_sample(X_train, y_train) lr_tl = LogisticRegression(max_iter=1000, class_weight='balanced') lr_tl.fit(X_train_tl, y_train_tl) y_pred_tl = lr_tl.predict(X_test) plot_confusion_matrix(y_test, y_pred_tl) # /SMOTE(Synthetic Minority OverSampling Technique)/ from imblearn.over_sampling import SMOTE smote = SMOTE(sampling_strategy='minority') X_train_sm, y_train_sm = smote.fit_sample(X_train, y_train) lr_sm = LogisticRegression(max_iter=1000) lr_sm.fit(X_train_sm, y_train_sm) y_pred_sm = lr_sm.predict(X_test) plot_confusion_matrix(y_test, y_pred_sm)
#
# manifold.manifold(imgs_origin, labels_origin, manifold_args, scores_predict,
# index_to_class=index_to_class, showLabels=False, showImages=False,
# imageZoom=0.15, imageDist=8e-3)
# %%
from imblearn.combine import SMOTEENN, SMOTETomek
from imblearn.under_sampling import RandomUnderSampler, TomekLinks
from collections import Counter
print(sorted(Counter(labels_origin).items()))
smote_enn = TomekLinks(random_state=0)
scores_predict2, labels_origin2 = smote_enn.fit_sample(
scores_predict, labels_origin)
print(sorted(Counter(labels_origin2).items()))
# %%
import manifold
manifold_args = dict(
RandomTrees=True,
PCA=True,
LinearDiscriminant=True,
Spectral=True,
TSNE=True,
n_neighbors=30)
index_to_class = io.reverse_dict(class_to_index)
def test_deprecation_random_state():
tl = TomekLinks(random_state=0)
with warns(DeprecationWarning,
match="'random_state' is deprecated from 0.4"):
tl.fit_sample(X, Y)
nm1 = NearMiss(random_state=0, version=1) X_resampled_nm1, y_resampled = nm1.fit_sample(X, y) print(sorted(Counter(y_resampled).items())) ''' Cleaning under-sampling techniques omek’s links TomekLinks:样本x与样本y来自于不同的类别,满足以下条件,它们之间被称之为TomekLinks; 不存在另外一个样本z,使得d(x,z)<d(x,y)或者 d(y,z)<d(x,y)成立.其中d(.)表示两个样本之间的距离,也就是说两个样本之间互为近邻关系. 这个时候,样本x或样本y很有可能是噪声数据,或者两个样本在边界的位置附近. TomekLinks函数中的auto参数控制Tomek’s links中的哪些样本被剔除. 默认的ratio='auto'移除多数类的样本,当ratio='all'时,两个样本均被移除. ''' from imblearn.under_sampling import TomekLinks tl =TomekLinks(random_state=0,ratio='all') X_resampled, y_resampled = tl.fit_sample(X, y) print(sorted(Counter(y_resampled).items())) ''' [(0, 55), (1, 249), (2, 4654)] ''' ''' Edited data set using nearest neighbours EditedNearestNeighbours这种方法应用最近邻算法来编辑(edit)数据集, 找出那些与邻居不太友好的样本然后移除.对于每一个要进行下采样的样本,那些不满足一些准则的样本将会被移除; 他们的绝大多数(kind_sel='mode')或者全部(kind_sel='all')的近邻样本都属于同一个类,这些样本会被保留在数据集中. ''' print(sorted(Counter(y).items())) from imblearn.under_sampling import EditedNearestNeighbours enn = EditedNearestNeighbours(random_state=0) X_resampled, y_resampled = enn.fit_sample(X, y) print(sorted(Counter(y_resampled).items()))
X_ros, y_ros = ros.fit_sample(X, y)
print(X_ros.shape[0] - X.shape[0], 'new random picked points')
plot_2d_space(X_ros, y_ros, 'Random over-sampling')
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(return_indices=True, ratio='majority')
X_tl, y_tl, id_tl = tl.fit_sample(X, y)
print('Removed indexes:', id_tl)
plot_2d_space(X_tl, y_tl, 'Tomek links under-sampling')
from imblearn.under_sampling import ClusterCentroids
cc = ClusterCentroids(ratio={0: 10})
X_cc, y_cc = cc.fit_sample(X, y)
# In[36]:
X_imb_train, X_imb_test, y_imb_train, y_imb_test = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0)
print(get_report(y_imb_train, y_imb_train))
# ### Under-Sampling with TomekLinks
# In[39]:
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(return_indices=False, ratio='majority')
X_tl, y_tl = tl.fit_sample(X_imb_train, y_imb_train)
plot_2d_space(X_tl, y_tl, 'Tomek links under-sampling')
# In[40]:
from sklearn.ensemble import RandomForestClassifier
sample_weights = np.array([1 if i == 0 else 5 for i in y_tl])
rfc_weighted_balanced = RandomForestClassifier(n_jobs=-1, warm_start=True)
rfc_weighted_balanced.fit(X_tl, y_tl, sample_weight=sample_weights)
print(get_report(y_imb_test, rfc_weighted_balanced.predict(X_imb_test)))
# Not good. lets try somthing else...
# from imblearn.under_sampling import NearMiss
# nm1 = NearMiss(random_state=0, version=1)
# X_resampled_nm1, y_resampled = nm1.fit_sample(X, y)
# print(sorted(Counter(y_resampled).items()))
'''
Cleaning under-sampling techniques
omek’s links
TomekLinks:样本x与样本y来自于不同的类别,满足以下条件,它们之间被称之为TomekLinks;
不存在另外一个样本z,使得d(x,z)<d(x,y)或者 d(y,z)<d(x,y)成立.其中d(.)表示两个样本之间的距离,也就是说两个样本之间互为近邻关系.
这个时候,样本x或样本y很有可能是噪声数据,或者两个样本在边界的位置附近.
TomekLinks函数中的auto参数控制Tomek’s links中的哪些样本被剔除.
默认的ratio='auto'移除多数类的样本,当ratio='all'时,两个样本均被移除.
'''
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(random_state=0, ratio='all')
X_resampled_tl, y_resampled_tl = tl.fit_sample(train_set_1_1, label)
print('TomekLinks ;', sorted(Counter(y_resampled_cc).items()))
x_train_tl, x_test_tl, y_train_tl, y_test_tl = train_test_split(X_resampled_tl,
y_resampled_tl,
random_state=1)
svm_clf.fit(x_train_tl, y_train_tl)
joblib.dump(svm_clf, '../model/tl_sample_model.pkl')
#tl评估
from sklearn.model_selection import cross_val_score
scores = cross_val_score(svm_clf, x_test_tl, y_test_tl, cv=5)
print('tl_score:', scores)
pred6 = svm_clf.predict(x_test_cc)
print('tl_accuracy_score:', metrics.accuracy_score(y_test_cc, pred3))
print('tl_f1_score:', metrics.f1_score(y_test_tl, pred6, average="micro"))
from sklearn.metrics import cohen_kappa_score #Kappa系数是基于混淆矩阵的计算得到的模型评价参数
print(__doc__)
rng = np.random.RandomState(0)
n_samples_1 = 500
n_samples_2 = 50
X_syn = np.r_[1.5 * rng.randn(n_samples_1, 2),
0.5 * rng.randn(n_samples_2, 2) + [2, 2]]
y_syn = np.array([0] * (n_samples_1) + [1] * (n_samples_2))
X_syn, y_syn = shuffle(X_syn, y_syn)
X_syn_train, X_syn_test, y_syn_train, y_syn_test = train_test_split(X_syn,
y_syn)
# remove Tomek links
tl = TomekLinks(return_indices=True)
X_resampled, y_resampled, idx_resampled = tl.fit_sample(X_syn, y_syn)
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
idx_samples_removed = np.setdiff1d(np.arange(X_syn.shape[0]),
idx_resampled)
idx_class_0 = y_resampled == 0
plt.scatter(X_resampled[idx_class_0, 0], X_resampled[idx_class_0, 1],
alpha=.8, label='Class #0')
plt.scatter(X_resampled[~idx_class_0, 0], X_resampled[~idx_class_0, 1],
alpha=.8, label='Class #1')
plt.scatter(X_syn[idx_samples_removed, 0], X_syn[idx_samples_removed, 1],
alpha=.8, label='Removed samples')
# make nice plotting
from imblearn.under_sampling import TomekLinks
# Generate the dataset
X, y = make_classification(n_classes=2, class_sep=2, weights=[0.1, 0.9],
n_informative=3, n_redundant=1, flip_y=0,
n_features=20, n_clusters_per_class=1,
n_samples=5000, random_state=10)
# Instanciate a PCA object for the sake of easy visualisation
pca = PCA(n_components=2)
# Fit and transform x to visualise inside a 2D feature space
X_vis = pca.fit_transform(X)
# Apply Tomek Links cleaning
tl = TomekLinks()
X_resampled, y_resampled = tl.fit_sample(X, y)
X_res_vis = pca.transform(X_resampled)
# Two subplots, unpack the axes array immediately
f, (ax1, ax2) = plt.subplots(1, 2)
ax1.scatter(X_vis[y == 0, 0], X_vis[y == 0, 1], label="Class #0", alpha=0.5,
edgecolor=almost_black, facecolor=palette[0], linewidth=0.15)
ax1.scatter(X_vis[y == 1, 0], X_vis[y == 1, 1], label="Class #1", alpha=0.5,
edgecolor=almost_black, facecolor=palette[2], linewidth=0.15)
ax1.set_title('Original set')
ax2.scatter(X_res_vis[y_resampled == 0, 0], X_res_vis[y_resampled == 0, 1],
label="Class #0", alpha=.5, edgecolor=almost_black,
facecolor=palette[0], linewidth=0.15)
ax2.scatter(X_res_vis[y_resampled == 1, 0], X_res_vis[y_resampled == 1, 1],
